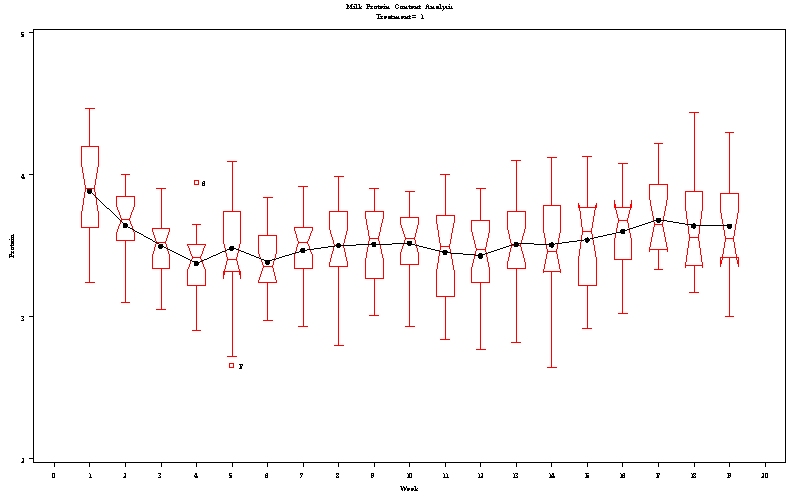
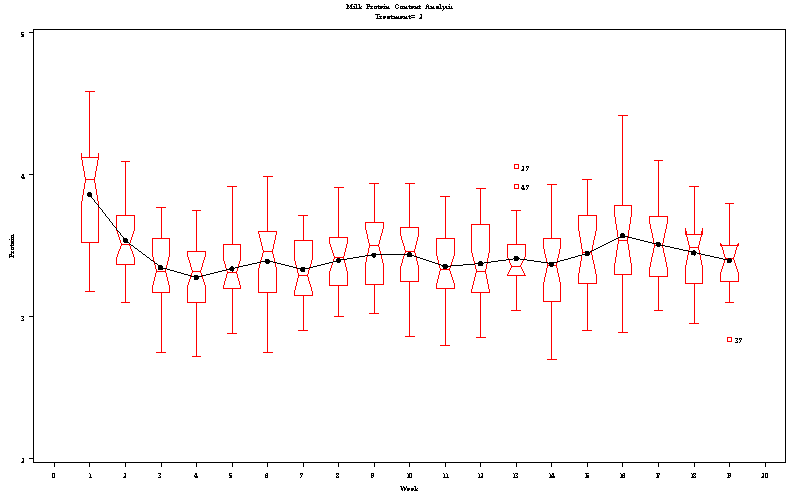
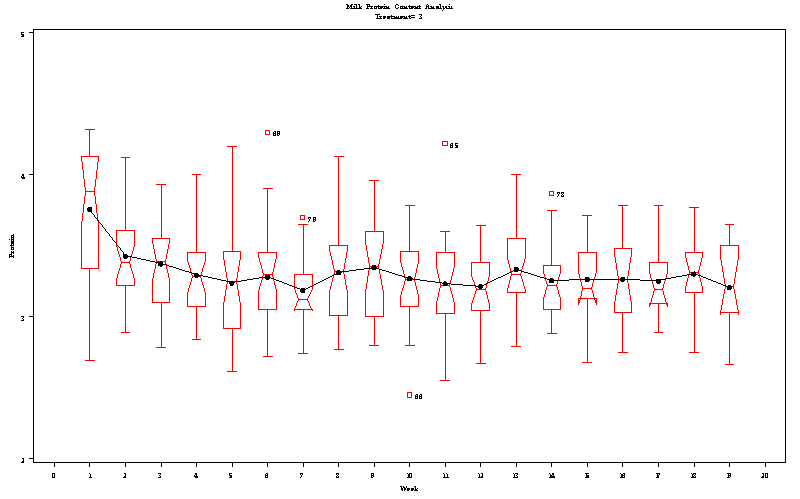
| Milk Protein Content Analysis |
| Multivariate Model |
| Model Information | |
|---|---|
| Data Set | WORK.MILK |
| Dependent Variable | Protein |
| Covariance Structure | Unstructured |
| Subject Effect | Cow(Treatment) |
| Estimation Method | REML |
| Residual Variance Method | None |
| Fixed Effects SE Method | Prasad-Rao-Jeske-Kackar-Harville |
| Degrees of Freedom Method | Kenward-Roger |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Cow | 79 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 |
| Treatment | 3 | 1 2 3 |
| Week | 19 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
| Dimensions | |
|---|---|
| Covariance Parameters | 190 |
| Columns in X | 80 |
| Columns in Z | 0 |
| Subjects | 79 |
| Max Obs Per Subject | 19 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 1501 |
| Number of Observations Used | 1337 |
| Number of Observations Not Used | 164 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 0 | 1 | 733.47680176 | |
| 1 | 2 | 586.83922651 | 13296908336 |
| 2 | 1 | 373.86373119 | 0.26179605 |
| 3 | 1 | 322.69564043 | 0.19479639 |
| 4 | 1 | 66.13778684 | 0.10685576 |
| 5 | 1 | -95.35447301 | 0.06150382 |
| 6 | 1 | -194.95930375 | 0.03633165 |
| 7 | 1 | -256.07343179 | 0.02116720 |
| 8 | 1 | -292.36148946 | 0.01174013 |
| 9 | 1 | -312.59927455 | 0.00591523 |
| 10 | 1 | -322.73177019 | 0.00252867 |
| 11 | 1 | -326.99217575 | 0.00086760 |
| 12 | 1 | -328.43239970 | 0.00026980 |
| 13 | 1 | -328.87620749 | 0.00008268 |
| 14 | 1 | -329.01165998 | 0.00001907 |
| 15 | 1 | -329.04126971 | 0.00000181 |
| 16 | 1 | -329.04387045 | 0.00000002 |
| 17 | 1 | -329.04390316 | 0.00000000 |
| Convergence criteria met. |
| Estimated R Matrix for Cow(Treatment) 1 1 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Row | Col1 | Col2 | Col3 | Col4 | Col5 | Col6 | Col7 | Col8 | Col9 | Col10 | Col11 | Col12 | Col13 | Col14 | Col15 | Col16 | Col17 | Col18 | Col19 |
| 1 | 0.1621 | 0.04831 | 0.04907 | 0.03288 | 0.04387 | 0.03025 | 0.02582 | 0.02804 | 0.02501 | 0.01618 | 0.007152 | 0.02462 | 0.02937 | 0.01017 | -0.00854 | 0.02225 | 0.01941 | 0.004659 | 0.03357 |
| 2 | 0.04831 | 0.07465 | 0.03439 | 0.03584 | 0.04287 | 0.02832 | 0.02743 | 0.02931 | 0.03227 | 0.01719 | 0.01892 | 0.006132 | 0.02545 | 0.01402 | 0.006996 | 0.03929 | 0.04579 | 0.01641 | 0.03299 |
| 3 | 0.04907 | 0.03439 | 0.06658 | 0.03940 | 0.04970 | 0.02999 | 0.02440 | 0.03529 | 0.03193 | 0.009953 | 0.01421 | 0.008366 | 0.01095 | 0.004287 | -0.00073 | 0.01011 | 0.02502 | 0.01803 | 0.03140 |
| 4 | 0.03288 | 0.03584 | 0.03940 | 0.06456 | 0.05832 | 0.04250 | 0.03684 | 0.05008 | 0.03731 | 0.01721 | 0.01509 | 0.003488 | 0.01494 | 0.003158 | -0.00439 | 0.01728 | 0.03614 | 0.03035 | 0.03464 |
| 5 | 0.04387 | 0.04287 | 0.04970 | 0.05832 | 0.1119 | 0.05234 | 0.05059 | 0.07140 | 0.05736 | 0.02208 | 0.01661 | 0.009470 | 0.006363 | -0.01499 | -0.03175 | 0.01239 | 0.02271 | 0.01168 | 0.05101 |
| 6 | 0.03025 | 0.02832 | 0.02999 | 0.04250 | 0.05234 | 0.08662 | 0.04228 | 0.05009 | 0.04255 | 0.02358 | -0.00190 | 0.003111 | 0.02266 | 0.006507 | -0.01875 | -0.00424 | 0.02327 | 0.01066 | 0.01867 |
| 7 | 0.02582 | 0.02743 | 0.02440 | 0.03684 | 0.05059 | 0.04228 | 0.05822 | 0.05215 | 0.04345 | 0.03526 | 0.02160 | 0.01218 | 0.02216 | 0.007344 | -0.01227 | -0.00624 | 0.01050 | 0.01091 | 0.01577 |
| 8 | 0.02804 | 0.02931 | 0.03529 | 0.05008 | 0.07140 | 0.05009 | 0.05215 | 0.09524 | 0.06275 | 0.04658 | 0.03596 | 0.02167 | 0.02796 | 0.009012 | -0.01574 | -0.00676 | 0.02228 | 0.007974 | 0.02609 |
| 9 | 0.02501 | 0.03227 | 0.03193 | 0.03731 | 0.05736 | 0.04255 | 0.04345 | 0.06275 | 0.08838 | 0.04611 | 0.03667 | 0.02078 | 0.02528 | 0.01642 | -0.01036 | -0.00138 | 0.01601 | 0.000559 | 0.01322 |
| 10 | 0.01618 | 0.01719 | 0.009953 | 0.01721 | 0.02208 | 0.02358 | 0.03526 | 0.04658 | 0.04611 | 0.07787 | 0.05144 | 0.03774 | 0.04093 | 0.04236 | 0.01247 | -0.00331 | 0.01717 | 0.008761 | 0.002616 |
| 11 | 0.007152 | 0.01892 | 0.01421 | 0.01509 | 0.01661 | -0.00190 | 0.02160 | 0.03596 | 0.03667 | 0.05144 | 0.09411 | 0.05291 | 0.04938 | 0.05571 | 0.05258 | 0.01049 | 0.02462 | 0.02130 | -0.00272 |
| 12 | 0.02462 | 0.006132 | 0.008366 | 0.003488 | 0.009470 | 0.003111 | 0.01218 | 0.02167 | 0.02078 | 0.03774 | 0.05291 | 0.08081 | 0.05921 | 0.06169 | 0.05370 | 0.02284 | 0.01049 | 0.01634 | 0.01788 |
| 13 | 0.02937 | 0.02545 | 0.01095 | 0.01494 | 0.006363 | 0.02266 | 0.02216 | 0.02796 | 0.02528 | 0.04093 | 0.04938 | 0.05921 | 0.08294 | 0.07614 | 0.06490 | 0.03117 | 0.02548 | 0.03425 | 0.01882 |
| 14 | 0.01017 | 0.01402 | 0.004287 | 0.003158 | -0.01499 | 0.006507 | 0.007344 | 0.009012 | 0.01642 | 0.04236 | 0.05571 | 0.06169 | 0.07614 | 0.1025 | 0.09242 | 0.02816 | 0.02205 | 0.04239 | 0.004249 |
| 15 | -0.00854 | 0.006996 | -0.00073 | -0.00439 | -0.03175 | -0.01875 | -0.01227 | -0.01574 | -0.01036 | 0.01247 | 0.05258 | 0.05370 | 0.06490 | 0.09242 | 0.1348 | 0.04643 | 0.01937 | 0.05525 | 0.009739 |
| 16 | 0.02225 | 0.03929 | 0.01011 | 0.01728 | 0.01239 | -0.00424 | -0.00624 | -0.00676 | -0.00138 | -0.00331 | 0.01049 | 0.02284 | 0.03117 | 0.02816 | 0.04643 | 0.09892 | 0.05845 | 0.05096 | 0.06479 |
| 17 | 0.01941 | 0.04579 | 0.02502 | 0.03614 | 0.02271 | 0.02327 | 0.01050 | 0.02228 | 0.01601 | 0.01717 | 0.02462 | 0.01049 | 0.02548 | 0.02205 | 0.01937 | 0.05845 | 0.08208 | 0.05165 | 0.05569 |
| 18 | 0.004659 | 0.01641 | 0.01803 | 0.03035 | 0.01168 | 0.01066 | 0.01091 | 0.007974 | 0.000559 | 0.008761 | 0.02130 | 0.01634 | 0.03425 | 0.04239 | 0.05525 | 0.05096 | 0.05165 | 0.08082 | 0.05632 |
| 19 | 0.03357 | 0.03299 | 0.03140 | 0.03464 | 0.05101 | 0.01867 | 0.01577 | 0.02609 | 0.01322 | 0.002616 | -0.00272 | 0.01788 | 0.01882 | 0.004249 | 0.009739 | 0.06479 | 0.05569 | 0.05632 | 0.09921 |
| Estimated R Correlation Matrix for Cow(Treatment) 1 1 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Row | Col1 | Col2 | Col3 | Col4 | Col5 | Col6 | Col7 | Col8 | Col9 | Col10 | Col11 | Col12 | Col13 | Col14 | Col15 | Col16 | Col17 | Col18 | Col19 |
| 1 | 1.0000 | 0.4392 | 0.4724 | 0.3214 | 0.3257 | 0.2553 | 0.2658 | 0.2256 | 0.2090 | 0.1440 | 0.05790 | 0.2151 | 0.2533 | 0.07892 | -0.05774 | 0.1757 | 0.1683 | 0.04071 | 0.2647 |
| 2 | 0.4392 | 1.0000 | 0.4878 | 0.5163 | 0.4691 | 0.3522 | 0.4161 | 0.3476 | 0.3973 | 0.2255 | 0.2257 | 0.07896 | 0.3235 | 0.1603 | 0.06973 | 0.4572 | 0.5850 | 0.2112 | 0.3833 |
| 3 | 0.4724 | 0.4878 | 1.0000 | 0.6010 | 0.5758 | 0.3949 | 0.3919 | 0.4432 | 0.4163 | 0.1382 | 0.1795 | 0.1141 | 0.1474 | 0.05189 | -0.00774 | 0.1245 | 0.3384 | 0.2458 | 0.3864 |
| 4 | 0.3214 | 0.5163 | 0.6010 | 1.0000 | 0.6862 | 0.5683 | 0.6008 | 0.6386 | 0.4939 | 0.2427 | 0.1936 | 0.04829 | 0.2041 | 0.03882 | -0.04706 | 0.2163 | 0.4965 | 0.4201 | 0.4328 |
| 5 | 0.3257 | 0.4691 | 0.5758 | 0.6862 | 1.0000 | 0.5316 | 0.6268 | 0.6916 | 0.5768 | 0.2365 | 0.1618 | 0.09959 | 0.06604 | -0.1399 | -0.2585 | 0.1177 | 0.2370 | 0.1228 | 0.4841 |
| 6 | 0.2553 | 0.3522 | 0.3949 | 0.5683 | 0.5316 | 1.0000 | 0.5953 | 0.5514 | 0.4863 | 0.2871 | -0.02105 | 0.03718 | 0.2673 | 0.06905 | -0.1735 | -0.04585 | 0.2760 | 0.1274 | 0.2014 |
| 7 | 0.2658 | 0.4161 | 0.3919 | 0.6008 | 0.6268 | 0.5953 | 1.0000 | 0.7003 | 0.6057 | 0.5237 | 0.2918 | 0.1776 | 0.3190 | 0.09506 | -0.1385 | -0.08227 | 0.1519 | 0.1590 | 0.2075 |
| 8 | 0.2256 | 0.3476 | 0.4432 | 0.6386 | 0.6916 | 0.5514 | 0.7003 | 1.0000 | 0.6840 | 0.5409 | 0.3798 | 0.2470 | 0.3146 | 0.09121 | -0.1389 | -0.06960 | 0.2521 | 0.09089 | 0.2684 |
| 9 | 0.2090 | 0.3973 | 0.4163 | 0.4939 | 0.5768 | 0.4863 | 0.6057 | 0.6840 | 1.0000 | 0.5558 | 0.4021 | 0.2459 | 0.2953 | 0.1725 | -0.09492 | -0.01471 | 0.1880 | 0.006617 | 0.1411 |
| 10 | 0.1440 | 0.2255 | 0.1382 | 0.2427 | 0.2365 | 0.2871 | 0.5237 | 0.5409 | 0.5558 | 1.0000 | 0.6009 | 0.4757 | 0.5093 | 0.4742 | 0.1217 | -0.03777 | 0.2148 | 0.1104 | 0.02976 |
| 11 | 0.05790 | 0.2257 | 0.1795 | 0.1936 | 0.1618 | -0.02105 | 0.2918 | 0.3798 | 0.4021 | 0.6009 | 1.0000 | 0.6067 | 0.5589 | 0.5672 | 0.4667 | 0.1087 | 0.2801 | 0.2442 | -0.02816 |
| 12 | 0.2151 | 0.07896 | 0.1141 | 0.04829 | 0.09959 | 0.03718 | 0.1776 | 0.2470 | 0.2459 | 0.4757 | 0.6067 | 1.0000 | 0.7233 | 0.6778 | 0.5144 | 0.2554 | 0.1288 | 0.2022 | 0.1997 |
| 13 | 0.2533 | 0.3235 | 0.1474 | 0.2041 | 0.06604 | 0.2673 | 0.3190 | 0.3146 | 0.2953 | 0.5093 | 0.5589 | 0.7233 | 1.0000 | 0.8257 | 0.6137 | 0.3441 | 0.3088 | 0.4183 | 0.2074 |
| 14 | 0.07892 | 0.1603 | 0.05189 | 0.03882 | -0.1399 | 0.06905 | 0.09506 | 0.09121 | 0.1725 | 0.4742 | 0.5672 | 0.6778 | 0.8257 | 1.0000 | 0.7861 | 0.2797 | 0.2404 | 0.4657 | 0.04214 |
| 15 | -0.05774 | 0.06973 | -0.00774 | -0.04706 | -0.2585 | -0.1735 | -0.1385 | -0.1389 | -0.09492 | 0.1217 | 0.4667 | 0.5144 | 0.6137 | 0.7861 | 1.0000 | 0.4021 | 0.1841 | 0.5293 | 0.08420 |
| 16 | 0.1757 | 0.4572 | 0.1245 | 0.2163 | 0.1177 | -0.04585 | -0.08227 | -0.06960 | -0.01471 | -0.03777 | 0.1087 | 0.2554 | 0.3441 | 0.2797 | 0.4021 | 1.0000 | 0.6487 | 0.5699 | 0.6540 |
| 17 | 0.1683 | 0.5850 | 0.3384 | 0.4965 | 0.2370 | 0.2760 | 0.1519 | 0.2521 | 0.1880 | 0.2148 | 0.2801 | 0.1288 | 0.3088 | 0.2404 | 0.1841 | 0.6487 | 1.0000 | 0.6341 | 0.6172 |
| 18 | 0.04071 | 0.2112 | 0.2458 | 0.4201 | 0.1228 | 0.1274 | 0.1590 | 0.09089 | 0.006617 | 0.1104 | 0.2442 | 0.2022 | 0.4183 | 0.4657 | 0.5293 | 0.5699 | 0.6341 | 1.0000 | 0.6290 |
| 19 | 0.2647 | 0.3833 | 0.3864 | 0.4328 | 0.4841 | 0.2014 | 0.2075 | 0.2684 | 0.1411 | 0.02976 | -0.02816 | 0.1997 | 0.2074 | 0.04214 | 0.08420 | 0.6540 | 0.6172 | 0.6290 | 1.0000 |
| Covariance Parameter Estimates | ||
|---|---|---|
| Cov Parm | Subject | Estimate |
| UN(1,1) | Cow(Treatment) | 0.1621 |
| UN(2,1) | Cow(Treatment) | 0.04831 |
| UN(2,2) | Cow(Treatment) | 0.07465 |
| UN(3,1) | Cow(Treatment) | 0.04907 |
| UN(3,2) | Cow(Treatment) | 0.03439 |
| UN(3,3) | Cow(Treatment) | 0.06658 |
| UN(4,1) | Cow(Treatment) | 0.03288 |
| UN(4,2) | Cow(Treatment) | 0.03584 |
| UN(4,3) | Cow(Treatment) | 0.03940 |
| UN(4,4) | Cow(Treatment) | 0.06456 |
| UN(5,1) | Cow(Treatment) | 0.04387 |
| UN(5,2) | Cow(Treatment) | 0.04287 |
| UN(5,3) | Cow(Treatment) | 0.04970 |
| UN(5,4) | Cow(Treatment) | 0.05832 |
| UN(5,5) | Cow(Treatment) | 0.1119 |
| UN(6,1) | Cow(Treatment) | 0.03025 |
| UN(6,2) | Cow(Treatment) | 0.02832 |
| UN(6,3) | Cow(Treatment) | 0.02999 |
| UN(6,4) | Cow(Treatment) | 0.04250 |
| UN(6,5) | Cow(Treatment) | 0.05234 |
| UN(6,6) | Cow(Treatment) | 0.08662 |
| UN(7,1) | Cow(Treatment) | 0.02582 |
| UN(7,2) | Cow(Treatment) | 0.02743 |
| UN(7,3) | Cow(Treatment) | 0.02440 |
| UN(7,4) | Cow(Treatment) | 0.03684 |
| UN(7,5) | Cow(Treatment) | 0.05059 |
| UN(7,6) | Cow(Treatment) | 0.04228 |
| UN(7,7) | Cow(Treatment) | 0.05822 |
| UN(8,1) | Cow(Treatment) | 0.02804 |
| UN(8,2) | Cow(Treatment) | 0.02931 |
| UN(8,3) | Cow(Treatment) | 0.03529 |
| UN(8,4) | Cow(Treatment) | 0.05008 |
| UN(8,5) | Cow(Treatment) | 0.07140 |
| UN(8,6) | Cow(Treatment) | 0.05009 |
| UN(8,7) | Cow(Treatment) | 0.05215 |
| UN(8,8) | Cow(Treatment) | 0.09524 |
| UN(9,1) | Cow(Treatment) | 0.02501 |
| UN(9,2) | Cow(Treatment) | 0.03227 |
| UN(9,3) | Cow(Treatment) | 0.03193 |
| UN(9,4) | Cow(Treatment) | 0.03731 |
| UN(9,5) | Cow(Treatment) | 0.05736 |
| UN(9,6) | Cow(Treatment) | 0.04255 |
| UN(9,7) | Cow(Treatment) | 0.04345 |
| UN(9,8) | Cow(Treatment) | 0.06275 |
| UN(9,9) | Cow(Treatment) | 0.08838 |
| UN(10,1) | Cow(Treatment) | 0.01618 |
| UN(10,2) | Cow(Treatment) | 0.01719 |
| UN(10,3) | Cow(Treatment) | 0.009953 |
| UN(10,4) | Cow(Treatment) | 0.01721 |
| UN(10,5) | Cow(Treatment) | 0.02208 |
| UN(10,6) | Cow(Treatment) | 0.02358 |
| UN(10,7) | Cow(Treatment) | 0.03526 |
| UN(10,8) | Cow(Treatment) | 0.04658 |
| UN(10,9) | Cow(Treatment) | 0.04611 |
| UN(10,10) | Cow(Treatment) | 0.07787 |
| UN(11,1) | Cow(Treatment) | 0.007152 |
| UN(11,2) | Cow(Treatment) | 0.01892 |
| UN(11,3) | Cow(Treatment) | 0.01421 |
| UN(11,4) | Cow(Treatment) | 0.01509 |
| UN(11,5) | Cow(Treatment) | 0.01661 |
| UN(11,6) | Cow(Treatment) | -0.00190 |
| UN(11,7) | Cow(Treatment) | 0.02160 |
| UN(11,8) | Cow(Treatment) | 0.03596 |
| UN(11,9) | Cow(Treatment) | 0.03667 |
| UN(11,10) | Cow(Treatment) | 0.05144 |
| UN(11,11) | Cow(Treatment) | 0.09411 |
| UN(12,1) | Cow(Treatment) | 0.02462 |
| UN(12,2) | Cow(Treatment) | 0.006132 |
| UN(12,3) | Cow(Treatment) | 0.008366 |
| UN(12,4) | Cow(Treatment) | 0.003488 |
| UN(12,5) | Cow(Treatment) | 0.009470 |
| UN(12,6) | Cow(Treatment) | 0.003111 |
| UN(12,7) | Cow(Treatment) | 0.01218 |
| UN(12,8) | Cow(Treatment) | 0.02167 |
| UN(12,9) | Cow(Treatment) | 0.02078 |
| UN(12,10) | Cow(Treatment) | 0.03774 |
| UN(12,11) | Cow(Treatment) | 0.05291 |
| UN(12,12) | Cow(Treatment) | 0.08081 |
| UN(13,1) | Cow(Treatment) | 0.02937 |
| UN(13,2) | Cow(Treatment) | 0.02545 |
| UN(13,3) | Cow(Treatment) | 0.01095 |
| UN(13,4) | Cow(Treatment) | 0.01494 |
| UN(13,5) | Cow(Treatment) | 0.006363 |
| UN(13,6) | Cow(Treatment) | 0.02266 |
| UN(13,7) | Cow(Treatment) | 0.02216 |
| UN(13,8) | Cow(Treatment) | 0.02796 |
| UN(13,9) | Cow(Treatment) | 0.02528 |
| UN(13,10) | Cow(Treatment) | 0.04093 |
| UN(13,11) | Cow(Treatment) | 0.04938 |
| UN(13,12) | Cow(Treatment) | 0.05921 |
| UN(13,13) | Cow(Treatment) | 0.08294 |
| UN(14,1) | Cow(Treatment) | 0.01017 |
| UN(14,2) | Cow(Treatment) | 0.01402 |
| UN(14,3) | Cow(Treatment) | 0.004287 |
| UN(14,4) | Cow(Treatment) | 0.003158 |
| UN(14,5) | Cow(Treatment) | -0.01499 |
| UN(14,6) | Cow(Treatment) | 0.006507 |
| UN(14,7) | Cow(Treatment) | 0.007344 |
| UN(14,8) | Cow(Treatment) | 0.009012 |
| UN(14,9) | Cow(Treatment) | 0.01642 |
| UN(14,10) | Cow(Treatment) | 0.04236 |
| UN(14,11) | Cow(Treatment) | 0.05571 |
| UN(14,12) | Cow(Treatment) | 0.06169 |
| UN(14,13) | Cow(Treatment) | 0.07614 |
| UN(14,14) | Cow(Treatment) | 0.1025 |
| UN(15,1) | Cow(Treatment) | -0.00854 |
| UN(15,2) | Cow(Treatment) | 0.006996 |
| UN(15,3) | Cow(Treatment) | -0.00073 |
| UN(15,4) | Cow(Treatment) | -0.00439 |
| UN(15,5) | Cow(Treatment) | -0.03175 |
| UN(15,6) | Cow(Treatment) | -0.01875 |
| UN(15,7) | Cow(Treatment) | -0.01227 |
| UN(15,8) | Cow(Treatment) | -0.01574 |
| UN(15,9) | Cow(Treatment) | -0.01036 |
| UN(15,10) | Cow(Treatment) | 0.01247 |
| UN(15,11) | Cow(Treatment) | 0.05258 |
| UN(15,12) | Cow(Treatment) | 0.05370 |
| UN(15,13) | Cow(Treatment) | 0.06490 |
| UN(15,14) | Cow(Treatment) | 0.09242 |
| UN(15,15) | Cow(Treatment) | 0.1348 |
| UN(16,1) | Cow(Treatment) | 0.02225 |
| UN(16,2) | Cow(Treatment) | 0.03929 |
| UN(16,3) | Cow(Treatment) | 0.01011 |
| UN(16,4) | Cow(Treatment) | 0.01728 |
| UN(16,5) | Cow(Treatment) | 0.01239 |
| UN(16,6) | Cow(Treatment) | -0.00424 |
| UN(16,7) | Cow(Treatment) | -0.00624 |
| UN(16,8) | Cow(Treatment) | -0.00676 |
| UN(16,9) | Cow(Treatment) | -0.00138 |
| UN(16,10) | Cow(Treatment) | -0.00331 |
| UN(16,11) | Cow(Treatment) | 0.01049 |
| UN(16,12) | Cow(Treatment) | 0.02284 |
| UN(16,13) | Cow(Treatment) | 0.03117 |
| UN(16,14) | Cow(Treatment) | 0.02816 |
| UN(16,15) | Cow(Treatment) | 0.04643 |
| UN(16,16) | Cow(Treatment) | 0.09892 |
| UN(17,1) | Cow(Treatment) | 0.01941 |
| UN(17,2) | Cow(Treatment) | 0.04579 |
| UN(17,3) | Cow(Treatment) | 0.02502 |
| UN(17,4) | Cow(Treatment) | 0.03614 |
| UN(17,5) | Cow(Treatment) | 0.02271 |
| UN(17,6) | Cow(Treatment) | 0.02327 |
| UN(17,7) | Cow(Treatment) | 0.01050 |
| UN(17,8) | Cow(Treatment) | 0.02228 |
| UN(17,9) | Cow(Treatment) | 0.01601 |
| UN(17,10) | Cow(Treatment) | 0.01717 |
| UN(17,11) | Cow(Treatment) | 0.02462 |
| UN(17,12) | Cow(Treatment) | 0.01049 |
| UN(17,13) | Cow(Treatment) | 0.02548 |
| UN(17,14) | Cow(Treatment) | 0.02205 |
| UN(17,15) | Cow(Treatment) | 0.01937 |
| UN(17,16) | Cow(Treatment) | 0.05845 |
| UN(17,17) | Cow(Treatment) | 0.08208 |
| UN(18,1) | Cow(Treatment) | 0.004659 |
| UN(18,2) | Cow(Treatment) | 0.01641 |
| UN(18,3) | Cow(Treatment) | 0.01803 |
| UN(18,4) | Cow(Treatment) | 0.03035 |
| UN(18,5) | Cow(Treatment) | 0.01168 |
| UN(18,6) | Cow(Treatment) | 0.01066 |
| UN(18,7) | Cow(Treatment) | 0.01091 |
| UN(18,8) | Cow(Treatment) | 0.007974 |
| UN(18,9) | Cow(Treatment) | 0.000559 |
| UN(18,10) | Cow(Treatment) | 0.008761 |
| UN(18,11) | Cow(Treatment) | 0.02130 |
| UN(18,12) | Cow(Treatment) | 0.01634 |
| UN(18,13) | Cow(Treatment) | 0.03425 |
| UN(18,14) | Cow(Treatment) | 0.04239 |
| UN(18,15) | Cow(Treatment) | 0.05525 |
| UN(18,16) | Cow(Treatment) | 0.05096 |
| UN(18,17) | Cow(Treatment) | 0.05165 |
| UN(18,18) | Cow(Treatment) | 0.08082 |
| UN(19,1) | Cow(Treatment) | 0.03357 |
| UN(19,2) | Cow(Treatment) | 0.03299 |
| UN(19,3) | Cow(Treatment) | 0.03140 |
| UN(19,4) | Cow(Treatment) | 0.03464 |
| UN(19,5) | Cow(Treatment) | 0.05101 |
| UN(19,6) | Cow(Treatment) | 0.01867 |
| UN(19,7) | Cow(Treatment) | 0.01577 |
| UN(19,8) | Cow(Treatment) | 0.02609 |
| UN(19,9) | Cow(Treatment) | 0.01322 |
| UN(19,10) | Cow(Treatment) | 0.002616 |
| UN(19,11) | Cow(Treatment) | -0.00272 |
| UN(19,12) | Cow(Treatment) | 0.01788 |
| UN(19,13) | Cow(Treatment) | 0.01882 |
| UN(19,14) | Cow(Treatment) | 0.004249 |
| UN(19,15) | Cow(Treatment) | 0.009739 |
| UN(19,16) | Cow(Treatment) | 0.06479 |
| UN(19,17) | Cow(Treatment) | 0.05569 |
| UN(19,18) | Cow(Treatment) | 0.05632 |
| UN(19,19) | Cow(Treatment) | 0.09921 |
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | -329.0 |
| AIC (smaller is better) | 51.0 |
| AICC (smaller is better) | 117.6 |
| BIC (smaller is better) | 501.2 |
| Null Model Likelihood Ratio Test | ||
|---|---|---|
| DF | Chi-Square | Pr > ChiSq |
| 189 | 1062.52 | <.0001 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| Treatment | 2 | 70.7 | 10.22 | 0.0001 |
| Week | 18 | 35.7 | 8.66 | <.0001 |
| Treatment*Week | 36 | 61.5 | 0.80 | 0.7594 |
| Type 3 Hotelling-Lawley-McKeon Statistics | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| Week | 18 | 59 | 10.81 | <.0001 |
| Treatment*Week | 36 | 102 | 1.07 | 0.3819 |
| Contrasts | ||||
|---|---|---|---|---|
| Label | Num DF | Den DF | F Value | Pr > F |
| Barley vs Barley+Lupins | 1 | 70.4 | 5.20 | 0.0257 |
| Lupins vs Barley+Lupins | 1 | 71 | 5.22 | 0.0253 |
| Barley vs Lupins | 1 | 70.7 | 20.43 | <.0001 |
| Milk Protein Content Analysis |
| Homogeneous Variance Compound Symmetry |
| Model Information | |
|---|---|
| Data Set | WORK.MILK |
| Dependent Variable | Protein |
| Covariance Structure | Compound Symmetry |
| Subject Effect | Cow(Treatment) |
| Estimation Method | REML |
| Residual Variance Method | Profile |
| Fixed Effects SE Method | Prasad-Rao-Jeske-Kackar-Harville |
| Degrees of Freedom Method | Kenward-Roger |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Cow | 79 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 |
| Treatment | 3 | 1 2 3 |
| Week | 19 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
| Dimensions | |
|---|---|
| Covariance Parameters | 2 |
| Columns in X | 80 |
| Columns in Z | 0 |
| Subjects | 79 |
| Max Obs Per Subject | 19 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 1501 |
| Number of Observations Used | 1337 |
| Number of Observations Not Used | 164 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 0 | 1 | 733.47680176 | |
| 1 | 2 | 427.52217160 | 0.00000104 |
| 2 | 1 | 427.52116565 | 0.00000000 |
| Convergence criteria met. |
| Covariance Parameter Estimates | ||
|---|---|---|
| Cov Parm | Subject | Estimate |
| CS | Cow(Treatment) | 0.02833 |
| Residual | 0.06261 | |
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | 427.5 |
| AIC (smaller is better) | 431.5 |
| AICC (smaller is better) | 431.5 |
| BIC (smaller is better) | 436.3 |
| Null Model Likelihood Ratio Test | ||
|---|---|---|
| DF | Chi-Square | Pr > ChiSq |
| 1 | 305.96 | <.0001 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| Treatment | 2 | 76.7 | 9.19 | 0.0003 |
| Week | 18 | 1206 | 16.33 | <.0001 |
| Treatment*Week | 36 | 1206 | 0.67 | 0.9360 |
| Contrasts | ||||
|---|---|---|---|---|
| Label | Num DF | Den DF | F Value | Pr > F |
| Barley vs Barley+Lupins | 1 | 76.5 | 3.80 | 0.0548 |
| Lupins vs Barley+Lupins | 1 | 76.8 | 5.63 | 0.0201 |
| Barley vs Lupins | 1 | 76.7 | 18.29 | <.0001 |
| Milk Protein Content Analysis |
| Homogeneous Variance First-order Autoregressive |
| Model Information | |
|---|---|
| Data Set | WORK.MILK |
| Dependent Variable | Protein |
| Covariance Structures | Variance Components, Autoregressive |
| Subject Effect | Cow(Treatment) |
| Estimation Method | REML |
| Residual Variance Method | Profile |
| Fixed Effects SE Method | Prasad-Rao-Jeske-Kackar-Harville |
| Degrees of Freedom Method | Kenward-Roger |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Cow | 79 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 |
| Treatment | 3 | 1 2 3 |
| Week | 19 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
| Dimensions | |
|---|---|
| Covariance Parameters | 3 |
| Columns in X | 80 |
| Columns in Z | 79 |
| Subjects | 1 |
| Max Obs Per Subject | 1501 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 1501 |
| Number of Observations Used | 1337 |
| Number of Observations Not Used | 164 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 0 | 1 | 733.47680176 | |
| 1 | 3 | 126.92018363 | 0.00008740 |
| 2 | 1 | 126.83032515 | 0.00000099 |
| 3 | 1 | 126.82921389 | 0.00000000 |
| Convergence criteria met. |
| Covariance Parameter Estimates | ||
|---|---|---|
| Cov Parm | Subject | Estimate |
| Cow(Treatment) | 0.01838 | |
| AR(1) | Cow(Treatment) | 0.5646 |
| Residual | 0.07662 | |
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | 126.8 |
| AIC (smaller is better) | 132.8 |
| AICC (smaller is better) | 132.8 |
| BIC (smaller is better) | 139.9 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| Treatment | 2 | 77.4 | 9.12 | 0.0003 |
| Week | 18 | 1074 | 13.63 | <.0001 |
| Treatment*Week | 36 | 1120 | 0.84 | 0.7340 |
| Contrasts | ||||
|---|---|---|---|---|
| Label | Num DF | Den DF | F Value | Pr > F |
| Barley vs Barley+Lupins | 1 | 77 | 4.06 | 0.0475 |
| Lupins vs Barley+Lupins | 1 | 77.7 | 5.27 | 0.0244 |
| Barley vs Lupins | 1 | 77.4 | 18.19 | <.0001 |
| Milk Protein Content Analysis |
| Homogeneous Variance Toeplitz |
| Model Information | |
|---|---|
| Data Set | WORK.MILK |
| Dependent Variable | Protein |
| Covariance Structure | Toeplitz |
| Subject Effect | Cow(Treatment) |
| Estimation Method | REML |
| Residual Variance Method | Profile |
| Fixed Effects SE Method | Prasad-Rao-Jeske-Kackar-Harville |
| Degrees of Freedom Method | Kenward-Roger |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Cow | 79 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 |
| Treatment | 3 | 1 2 3 |
| Week | 19 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
| Dimensions | |
|---|---|
| Covariance Parameters | 19 |
| Columns in X | 80 |
| Columns in Z | 0 |
| Subjects | 79 |
| Max Obs Per Subject | 19 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 1501 |
| Number of Observations Used | 1337 |
| Number of Observations Not Used | 164 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 0 | 1 | 733.47680176 | |
| 1 | 2 | 47.11318133 | 0.01947966 |
| 2 | 1 | 20.44774678 | 0.00665614 |
| 3 | 1 | 11.44408368 | 0.00109281 |
| 4 | 1 | 10.01603821 | 0.00005429 |
| 5 | 1 | 9.95000492 | 0.00000020 |
| 6 | 1 | 9.94976484 | 0.00000000 |
| Convergence criteria met. |
| Covariance Parameter Estimates | ||
|---|---|---|
| Cov Parm | Subject | Estimate |
| TOEP(2) | Cow(Treatment) | 0.06216 |
| TOEP(3) | Cow(Treatment) | 0.05374 |
| TOEP(4) | Cow(Treatment) | 0.04807 |
| TOEP(5) | Cow(Treatment) | 0.03981 |
| TOEP(6) | Cow(Treatment) | 0.02696 |
| TOEP(7) | Cow(Treatment) | 0.01984 |
| TOEP(8) | Cow(Treatment) | 0.01745 |
| TOEP(9) | Cow(Treatment) | 0.009548 |
| TOEP(10) | Cow(Treatment) | 0.005451 |
| TOEP(11) | Cow(Treatment) | 0.004145 |
| TOEP(12) | Cow(Treatment) | 0.01370 |
| TOEP(13) | Cow(Treatment) | 0.01331 |
| TOEP(14) | Cow(Treatment) | 0.01379 |
| TOEP(15) | Cow(Treatment) | 0.02167 |
| TOEP(16) | Cow(Treatment) | 0.01840 |
| TOEP(17) | Cow(Treatment) | 0.01488 |
| TOEP(18) | Cow(Treatment) | 0.01211 |
| TOEP(19) | Cow(Treatment) | 0.01391 |
| Residual | 0.09562 | |
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | 9.9 |
| AIC (smaller is better) | 47.9 |
| AICC (smaller is better) | 48.6 |
| BIC (smaller is better) | 93.0 |
| Null Model Likelihood Ratio Test | ||
|---|---|---|
| DF | Chi-Square | Pr > ChiSq |
| 18 | 723.53 | <.0001 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| Treatment | 2 | 76 | 9.10 | 0.0003 |
| Week | 18 | 484 | 18.08 | <.0001 |
| Treatment*Week | 36 | 661 | 0.90 | 0.6400 |
| Contrasts | ||||
|---|---|---|---|---|
| Label | Num DF | Den DF | F Value | Pr > F |
| Barley vs Barley+Lupins | 1 | 75.7 | 4.28 | 0.0419 |
| Lupins vs Barley+Lupins | 1 | 76.2 | 5.00 | 0.0282 |
| Barley vs Lupins | 1 | 76 | 18.17 | <.0001 |
| Milk Protein Content Analysis |
| Heterogeneous Variance Compound Symmetry |
| Model Information | |
|---|---|
| Data Set | WORK.MILK |
| Dependent Variable | Protein |
| Covariance Structure | Heterogeneous Compound Symmetry |
| Subject Effect | Cow(Treatment) |
| Estimation Method | REML |
| Residual Variance Method | None |
| Fixed Effects SE Method | Prasad-Rao-Jeske-Kackar-Harville |
| Degrees of Freedom Method | Kenward-Roger |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Cow | 79 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 |
| Treatment | 3 | 1 2 3 |
| Week | 19 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
| Dimensions | |
|---|---|
| Covariance Parameters | 20 |
| Columns in X | 80 |
| Columns in Z | 0 |
| Subjects | 79 |
| Max Obs Per Subject | 19 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 1501 |
| Number of Observations Used | 1337 |
| Number of Observations Not Used | 164 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 0 | 1 | 733.47680176 | |
| 1 | 2 | 366.76734970 | 0.00011759 |
| 2 | 1 | 366.64593128 | 0.00000052 |
| 3 | 1 | 366.64541370 | 0.00000000 |
| Convergence criteria met. |
| Covariance Parameter Estimates | ||
|---|---|---|
| Cov Parm | Subject | Estimate |
| Var(1) | Cow(Treatment) | 0.1793 |
| Var(2) | Cow(Treatment) | 0.07249 |
| Var(3) | Cow(Treatment) | 0.06636 |
| Var(4) | Cow(Treatment) | 0.05950 |
| Var(5) | Cow(Treatment) | 0.1076 |
| Var(6) | Cow(Treatment) | 0.09005 |
| Var(7) | Cow(Treatment) | 0.05398 |
| Var(8) | Cow(Treatment) | 0.08647 |
| Var(9) | Cow(Treatment) | 0.08501 |
| Var(10) | Cow(Treatment) | 0.07689 |
| Var(11) | Cow(Treatment) | 0.09706 |
| Var(12) | Cow(Treatment) | 0.08553 |
| Var(13) | Cow(Treatment) | 0.08036 |
| Var(14) | Cow(Treatment) | 0.1107 |
| Var(15) | Cow(Treatment) | 0.1152 |
| Var(16) | Cow(Treatment) | 0.1265 |
| Var(17) | Cow(Treatment) | 0.08503 |
| Var(18) | Cow(Treatment) | 0.09110 |
| Var(19) | Cow(Treatment) | 0.1044 |
| CSH | Cow(Treatment) | 0.3294 |
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | 366.6 |
| AIC (smaller is better) | 406.6 |
| AICC (smaller is better) | 407.3 |
| BIC (smaller is better) | 454.0 |
| Null Model Likelihood Ratio Test | ||
|---|---|---|
| DF | Chi-Square | Pr > ChiSq |
| 19 | 366.83 | <.0001 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| Treatment | 2 | 76 | 8.52 | 0.0005 |
| Week | 18 | 461 | 10.81 | <.0001 |
| Treatment*Week | 36 | 630 | 0.72 | 0.8912 |
| Contrasts | ||||
|---|---|---|---|---|
| Label | Num DF | Den DF | F Value | Pr > F |
| Barley vs Barley+Lupins | 1 | 75.9 | 3.51 | 0.0650 |
| Lupins vs Barley+Lupins | 1 | 76.2 | 5.24 | 0.0248 |
| Barley vs Lupins | 1 | 76.1 | 16.95 | <.0001 |
| Milk Protein Content Analysis |
| Heterogeneous Variance First-order Autoregressive |
| Model Information | |
|---|---|
| Data Set | WORK.MILK |
| Dependent Variable | Protein |
| Covariance Structures | Variance Components, Heterogeneous Autoregressive |
| Subject Effect | Cow(Treatment) |
| Estimation Method | REML |
| Residual Variance Method | None |
| Fixed Effects SE Method | Prasad-Rao-Jeske-Kackar-Harville |
| Degrees of Freedom Method | Kenward-Roger |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Cow | 79 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 |
| Treatment | 3 | 1 2 3 |
| Week | 19 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
| Dimensions | |
|---|---|
| Covariance Parameters | 21 |
| Columns in X | 80 |
| Columns in Z | 79 |
| Subjects | 1 |
| Max Obs Per Subject | 1501 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 1501 |
| Number of Observations Used | 1337 |
| Number of Observations Not Used | 164 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 0 | 1 | 733.47680176 | |
| 1 | 2 | 51.32810427 | 0.00702647 |
| 2 | 1 | 41.73059104 | 0.00107803 |
| 3 | 1 | 40.33761697 | 0.00004744 |
| 4 | 1 | 40.28100325 | 0.00000012 |
| 5 | 1 | 40.28086190 | 0.00000000 |
| Convergence criteria met. |
| Covariance Parameter Estimates | ||
|---|---|---|
| Cov Parm | Subject | Estimate |
| Cow(Treatment) | 0.02192 | |
| Var(1) | Cow(Treatment) | 0.1773 |
| Var(2) | Cow(Treatment) | 0.07272 |
| Var(3) | Cow(Treatment) | 0.07017 |
| Var(4) | Cow(Treatment) | 0.04628 |
| Var(5) | Cow(Treatment) | 0.09874 |
| Var(6) | Cow(Treatment) | 0.07963 |
| Var(7) | Cow(Treatment) | 0.04430 |
| Var(8) | Cow(Treatment) | 0.06999 |
| Var(9) | Cow(Treatment) | 0.07623 |
| Var(10) | Cow(Treatment) | 0.06865 |
| Var(11) | Cow(Treatment) | 0.08127 |
| Var(12) | Cow(Treatment) | 0.06403 |
| Var(13) | Cow(Treatment) | 0.04270 |
| Var(14) | Cow(Treatment) | 0.06181 |
| Var(15) | Cow(Treatment) | 0.08725 |
| Var(16) | Cow(Treatment) | 0.09719 |
| Var(17) | Cow(Treatment) | 0.05388 |
| Var(18) | Cow(Treatment) | 0.05086 |
| Var(19) | Cow(Treatment) | 0.06662 |
| ARH(1) | Cow(Treatment) | 0.5625 |
| Fit Statistics | |
|---|---|
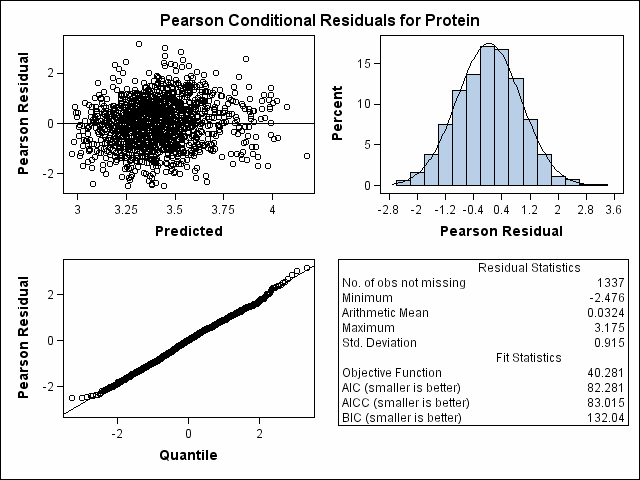
| -2 Res Log Likelihood | 40.3 |
| AIC (smaller is better) | 82.3 |
| AICC (smaller is better) | 83.0 |
| BIC (smaller is better) | 132.0 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| Treatment | 2 | 77.1 | 8.47 | 0.0005 |
| Week | 18 | 385 | 7.81 | <.0001 |
| Treatment*Week | 36 | 546 | 0.94 | 0.5766 |
| Contrasts | ||||
|---|---|---|---|---|
| Label | Num DF | Den DF | F Value | Pr > F |
| Barley vs Barley+Lupins | 1 | 76.9 | 3.72 | 0.0574 |
| Lupins vs Barley+Lupins | 1 | 77.4 | 4.94 | 0.0291 |
| Barley vs Lupins | 1 | 77.2 | 16.88 | <.0001 |
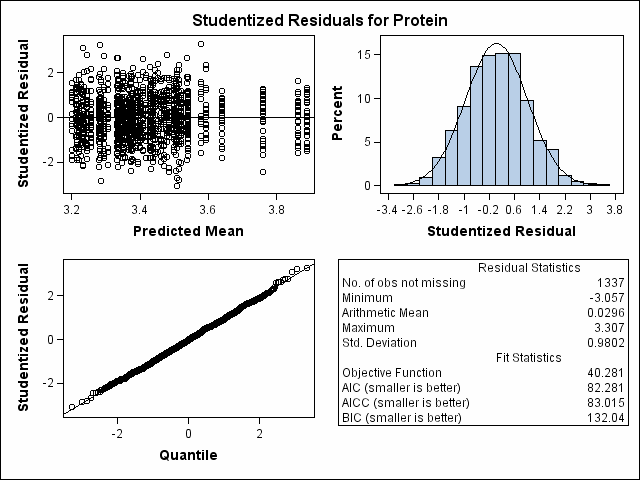
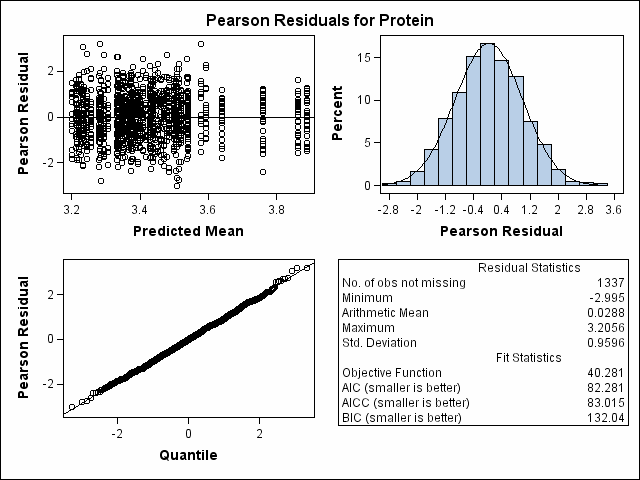
| Milk Protein Content Analysis |
| Heterogeneous Variance First-order Autoregressive |
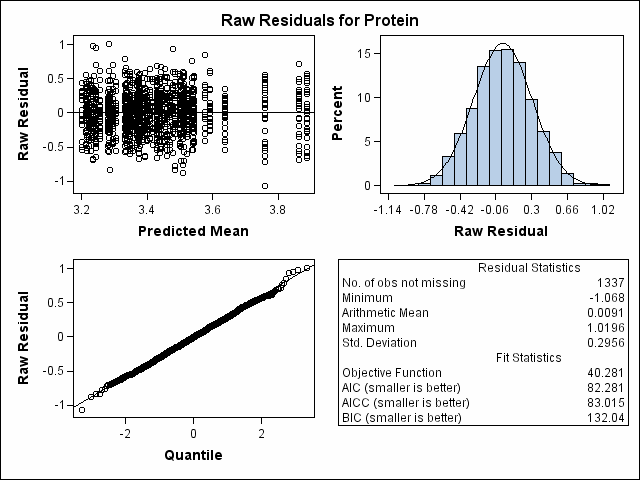
| Milk Protein Content Analysis |
| Heterogeneous Variance First-order Autoregressive |
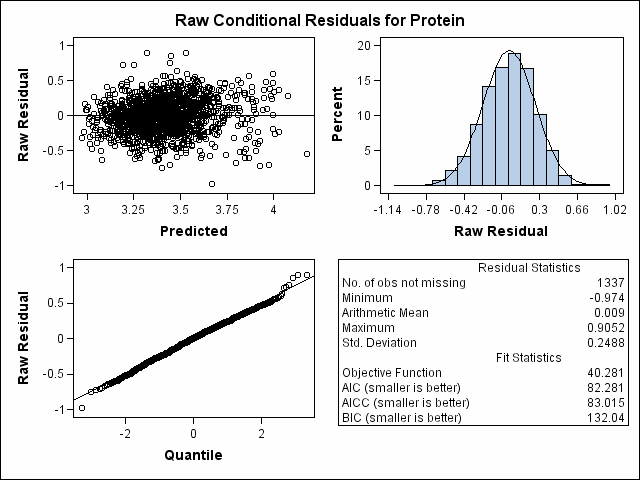
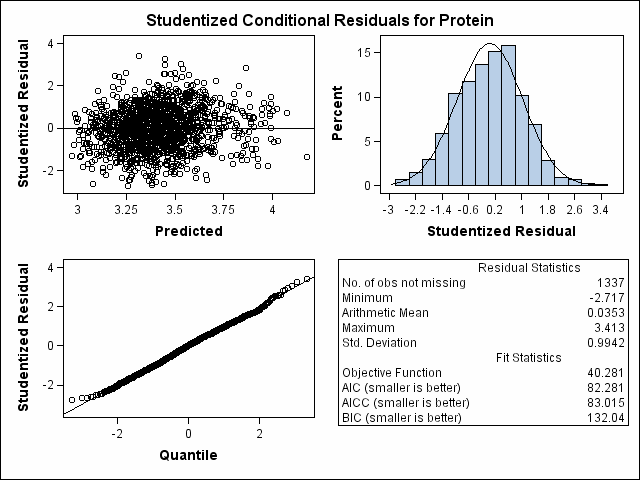
| Milk Protein Content Analysis |
| Heterogeneous Variance First-order Antedependence |
| Model Information | |
|---|---|
| Data Set | WORK.MILK |
| Dependent Variable | Protein |
| Covariance Structure | Ante-dependence |
| Subject Effect | Cow(Treatment) |
| Estimation Method | REML |
| Residual Variance Method | None |
| Fixed Effects SE Method | Prasad-Rao-Jeske-Kackar-Harville |
| Degrees of Freedom Method | Kenward-Roger |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Cow | 79 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 |
| Treatment | 3 | 1 2 3 |
| Week | 19 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
| Dimensions | |
|---|---|
| Covariance Parameters | 37 |
| Columns in X | 80 |
| Columns in Z | 0 |
| Subjects | 79 |
| Max Obs Per Subject | 19 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 1501 |
| Number of Observations Used | 1337 |
| Number of Observations Not Used | 164 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 0 | 1 | 733.47680176 | |
| 1 | 2 | 30.77089921 | 0.00299314 |
| 2 | 1 | 27.97598971 | 0.00031666 |
| 3 | 1 | 27.58223651 | 0.00000587 |
| 4 | 1 | 27.57528308 | 0.00000001 |
| Convergence criteria met. |
| Covariance Parameter Estimates | ||
|---|---|---|
| Cov Parm | Subject | Estimate |
| Var(1) | Cow(Treatment) | 0.1621 |
| Var(2) | Cow(Treatment) | 0.07489 |
| Var(3) | Cow(Treatment) | 0.06658 |
| Var(4) | Cow(Treatment) | 0.06456 |
| Var(5) | Cow(Treatment) | 0.1098 |
| Var(6) | Cow(Treatment) | 0.08662 |
| Var(7) | Cow(Treatment) | 0.06183 |
| Var(8) | Cow(Treatment) | 0.09821 |
| Var(9) | Cow(Treatment) | 0.08904 |
| Var(10) | Cow(Treatment) | 0.07784 |
| Var(11) | Cow(Treatment) | 0.09395 |
| Var(12) | Cow(Treatment) | 0.08081 |
| Var(13) | Cow(Treatment) | 0.08204 |
| Var(14) | Cow(Treatment) | 0.1025 |
| Var(15) | Cow(Treatment) | 0.1256 |
| Var(16) | Cow(Treatment) | 0.1354 |
| Var(17) | Cow(Treatment) | 0.09902 |
| Var(18) | Cow(Treatment) | 0.09862 |
| Var(19) | Cow(Treatment) | 0.1124 |
| Rho(1) | Cow(Treatment) | 0.4382 |
| Rho(2) | Cow(Treatment) | 0.4852 |
| Rho(3) | Cow(Treatment) | 0.6010 |
| Rho(4) | Cow(Treatment) | 0.6858 |
| Rho(5) | Cow(Treatment) | 0.5344 |
| Rho(6) | Cow(Treatment) | 0.6241 |
| Rho(7) | Cow(Treatment) | 0.7208 |
| Rho(8) | Cow(Treatment) | 0.6867 |
| Rho(9) | Cow(Treatment) | 0.5546 |
| Rho(10) | Cow(Treatment) | 0.5986 |
| Rho(11) | Cow(Treatment) | 0.6070 |
| Rho(12) | Cow(Treatment) | 0.7332 |
| Rho(13) | Cow(Treatment) | 0.8223 |
| Rho(14) | Cow(Treatment) | 0.7750 |
| Rho(15) | Cow(Treatment) | 0.6429 |
| Rho(16) | Cow(Treatment) | 0.7778 |
| Rho(17) | Cow(Treatment) | 0.7717 |
| Rho(18) | Cow(Treatment) | 0.8214 |
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | 27.6 |
| AIC (smaller is better) | 101.6 |
| AICC (smaller is better) | 103.8 |
| BIC (smaller is better) | 189.2 |
| Null Model Likelihood Ratio Test | ||
|---|---|---|
| DF | Chi-Square | Pr > ChiSq |
| 36 | 705.90 | <.0001 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| Treatment | 2 | 116 | 13.01 | <.0001 |
| Week | 18 | 265 | 8.07 | <.0001 |
| Treatment*Week | 36 | 407 | 0.92 | 0.6040 |
| Contrasts | ||||
|---|---|---|---|---|
| Label | Num DF | Den DF | F Value | Pr > F |
| Barley vs Barley+Lupins | 1 | 116 | 5.88 | 0.0169 |
| Lupins vs Barley+Lupins | 1 | 117 | 7.42 | 0.0074 |
| Barley vs Lupins | 1 | 117 | 25.96 | <.0001 |
| Milk Protein Content Analysis |
| Heterogeneous Variance Toeplitz |
| Model Information | |
|---|---|
| Data Set | WORK.MILK |
| Dependent Variable | Protein |
| Covariance Structure | Heterogeneous Toeplitz |
| Subject Effect | Cow(Treatment) |
| Estimation Method | REML |
| Residual Variance Method | None |
| Fixed Effects SE Method | Prasad-Rao-Jeske-Kackar-Harville |
| Degrees of Freedom Method | Kenward-Roger |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Cow | 79 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 |
| Treatment | 3 | 1 2 3 |
| Week | 19 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
| Dimensions | |
|---|---|
| Covariance Parameters | 37 |
| Columns in X | 80 |
| Columns in Z | 0 |
| Subjects | 79 |
| Max Obs Per Subject | 19 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 1501 |
| Number of Observations Used | 1337 |
| Number of Observations Not Used | 164 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 0 | 1 | 733.47680176 | |
| 1 | 2 | -49.93394814 | 0.01199920 |
| 2 | 1 | -66.73011736 | 0.00138150 |
| 3 | 1 | -68.57449245 | 0.00004763 |
| 4 | 1 | -68.63369972 | 0.00000009 |
| 5 | 1 | -68.63380358 | 0.00000000 |
| Convergence criteria met. |
| Covariance Parameter Estimates | ||
|---|---|---|
| Cov Parm | Subject | Estimate |
| Var(1) | Cow(Treatment) | 0.2189 |
| Var(2) | Cow(Treatment) | 0.09449 |
| Var(3) | Cow(Treatment) | 0.08635 |
| Var(4) | Cow(Treatment) | 0.06687 |
| Var(5) | Cow(Treatment) | 0.1086 |
| Var(6) | Cow(Treatment) | 0.08976 |
| Var(7) | Cow(Treatment) | 0.05748 |
| Var(8) | Cow(Treatment) | 0.09458 |
| Var(9) | Cow(Treatment) | 0.09710 |
| Var(10) | Cow(Treatment) | 0.08582 |
| Var(11) | Cow(Treatment) | 0.09757 |
| Var(12) | Cow(Treatment) | 0.08147 |
| Var(13) | Cow(Treatment) | 0.07202 |
| Var(14) | Cow(Treatment) | 0.08679 |
| Var(15) | Cow(Treatment) | 0.1002 |
| Var(16) | Cow(Treatment) | 0.1046 |
| Var(17) | Cow(Treatment) | 0.07530 |
| Var(18) | Cow(Treatment) | 0.06829 |
| Var(19) | Cow(Treatment) | 0.08169 |
| TOEPH(1) | Cow(Treatment) | 0.6499 |
| TOEPH(2) | Cow(Treatment) | 0.5580 |
| TOEPH(3) | Cow(Treatment) | 0.4936 |
| TOEPH(4) | Cow(Treatment) | 0.4000 |
| TOEPH(5) | Cow(Treatment) | 0.2709 |
| TOEPH(6) | Cow(Treatment) | 0.1966 |
| TOEPH(7) | Cow(Treatment) | 0.1676 |
| TOEPH(8) | Cow(Treatment) | 0.07914 |
| TOEPH(9) | Cow(Treatment) | 0.04599 |
| TOEPH(10) | Cow(Treatment) | 0.03811 |
| TOEPH(11) | Cow(Treatment) | 0.1462 |
| TOEPH(12) | Cow(Treatment) | 0.1401 |
| TOEPH(13) | Cow(Treatment) | 0.1675 |
| TOEPH(14) | Cow(Treatment) | 0.2838 |
| TOEPH(15) | Cow(Treatment) | 0.2286 |
| TOEPH(16) | Cow(Treatment) | 0.1816 |
| TOEPH(17) | Cow(Treatment) | 0.1348 |
| TOEPH(18) | Cow(Treatment) | 0.1444 |
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | -68.6 |
| AIC (smaller is better) | 5.4 |
| AICC (smaller is better) | 7.6 |
| BIC (smaller is better) | 93.0 |
| Null Model Likelihood Ratio Test | ||
|---|---|---|
| DF | Chi-Square | Pr > ChiSq |
| 36 | 802.11 | <.0001 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| Treatment | 2 | 76.5 | 9.79 | 0.0002 |
| Week | 18 | 276 | 10.51 | <.0001 |
| Treatment*Week | 36 | 420 | 1.00 | 0.4668 |
| Contrasts | ||||
|---|---|---|---|---|
| Label | Num DF | Den DF | F Value | Pr > F |
| Barley vs Barley+Lupins | 1 | 76.2 | 4.65 | 0.0343 |
| Lupins vs Barley+Lupins | 1 | 76.7 | 5.34 | 0.0235 |
| Barley vs Lupins | 1 | 76.5 | 19.55 | <.0001 |
| Milk Protein Content Analysis |
| Continuous Time Spatial Power Covariogram |
| Model Information | |
|---|---|
| Data Set | WORK.MILK |
| Dependent Variable | Protein |
| Covariance Structures | Variance Components, Spatial Power |
| Subject Effect | Cow(Treatment) |
| Estimation Method | REML |
| Residual Variance Method | Profile |
| Fixed Effects SE Method | Prasad-Rao-Jeske-Kackar-Harville |
| Degrees of Freedom Method | Kenward-Roger |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Cow | 79 | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 |
| Treatment | 3 | 1 2 3 |
| Dimensions | |
|---|---|
| Covariance Parameters | 3 |
| Columns in X | 17 |
| Columns in Z | 79 |
| Subjects | 1 |
| Max Obs Per Subject | 1501 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 1501 |
| Number of Observations Used | 1337 |
| Number of Observations Not Used | 164 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 0 | 1 | 726.79675947 | |
| 1 | 3 | 107.34044489 | 0.00010121 |
| 2 | 2 | 107.23283283 | 0.00000142 |
| 3 | 1 | 107.23116616 | 0.00000000 |
| Convergence criteria met. |
| Estimated R Correlation Matrix for Cow(Treatment) 1 1 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Row | Col1 | Col2 | Col3 | Col4 | Col5 | Col6 | Col7 | Col8 | Col9 | Col10 | Col11 | Col12 | Col13 | Col14 | Col15 | Col16 | Col17 | Col18 | Col19 |
| 1 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 | 0.006589 | 0.003771 | 0.002158 | 0.001235 | 0.000707 | 0.000405 | 0.000232 | 0.000133 | 0.000076 | 0.000043 |
| 2 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 | 0.006589 | 0.003771 | 0.002158 | 0.001235 | 0.000707 | 0.000405 | 0.000232 | 0.000133 | 0.000076 |
| 3 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 | 0.006589 | 0.003771 | 0.002158 | 0.001235 | 0.000707 | 0.000405 | 0.000232 | 0.000133 |
| 4 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 | 0.006589 | 0.003771 | 0.002158 | 0.001235 | 0.000707 | 0.000405 | 0.000232 |
| 5 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 | 0.006589 | 0.003771 | 0.002158 | 0.001235 | 0.000707 | 0.000405 |
| 6 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 | 0.006589 | 0.003771 | 0.002158 | 0.001235 | 0.000707 |
| 7 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 | 0.006589 | 0.003771 | 0.002158 | 0.001235 |
| 8 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 | 0.006589 | 0.003771 | 0.002158 |
| 9 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 | 0.006589 | 0.003771 |
| 10 | 0.006589 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 | 0.006589 |
| 11 | 0.003771 | 0.006589 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 | 0.01151 |
| 12 | 0.002158 | 0.003771 | 0.006589 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 | 0.02011 |
| 13 | 0.001235 | 0.002158 | 0.003771 | 0.006589 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 | 0.03514 |
| 14 | 0.000707 | 0.001235 | 0.002158 | 0.003771 | 0.006589 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 | 0.06141 |
| 15 | 0.000405 | 0.000707 | 0.001235 | 0.002158 | 0.003771 | 0.006589 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 | 0.1073 |
| 16 | 0.000232 | 0.000405 | 0.000707 | 0.001235 | 0.002158 | 0.003771 | 0.006589 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 | 0.1875 |
| 17 | 0.000133 | 0.000232 | 0.000405 | 0.000707 | 0.001235 | 0.002158 | 0.003771 | 0.006589 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 | 0.3276 |
| 18 | 0.000076 | 0.000133 | 0.000232 | 0.000405 | 0.000707 | 0.001235 | 0.002158 | 0.003771 | 0.006589 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 | 0.5723 |
| 19 | 0.000043 | 0.000076 | 0.000133 | 0.000232 | 0.000405 | 0.000707 | 0.001235 | 0.002158 | 0.003771 | 0.006589 | 0.01151 | 0.02011 | 0.03514 | 0.06141 | 0.1073 | 0.1875 | 0.3276 | 0.5723 | 1.0000 |
| Covariance Parameter Estimates | ||
|---|---|---|
| Cov Parm | Subject | Estimate |
| Cow(Treatment) | 0.01764 | |
| SP(POW) | Cow(Treatment) | 0.5723 |
| Residual | 0.07935 | |
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | 107.2 |
| AIC (smaller is better) | 113.2 |
| AICC (smaller is better) | 113.2 |
| BIC (smaller is better) | 120.3 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| Treatment | 2 | 575 | 0.76 | 0.4662 |
| Week | 1 | 858 | 111.73 | <.0001 |
| Week*Week | 1 | 773 | 66.43 | <.0001 |
| Week*Week*Week | 1 | 746 | 44.68 | <.0001 |
| Week*Week*Week*Week | 1 | 750 | 32.63 | <.0001 |
| Week*Treatment | 2 | 485 | 0.51 | 0.5981 |
| Week*Week*Treatment | 2 | 460 | 0.57 | 0.5632 |
| Week*Week*Week*Treat | 2 | 464 | 0.59 | 0.5534 |
| Contrasts | ||||
|---|---|---|---|---|
| Label | Num DF | Den DF | F Value | Pr > F |
| Barley vs Barley+Lupins | 1 | 575 | 0.00 | 0.9804 |
| Lupins vs Barley+Lupins | 1 | 575 | 1.19 | 0.2765 |
| Barley vs Lupins | 1 | 576 | 1.09 | 0.2971 |
| Least Squares Means | |||||||
|---|---|---|---|---|---|---|---|
| Effect | Treatment | Week | Estimate | Standard Error | DF | t Value | Pr > |t| |
| Treatment | 1 | 1.00 | 3.8789 | 0.06133 | 391 | 63.25 | <.0001 |
| Treatment | 2 | 1.00 | 3.8405 | 0.05903 | 391 | 65.06 | <.0001 |
| Treatment | 3 | 1.00 | 3.7459 | 0.05903 | 392 | 63.46 | <.0001 |
| Treatment | 1 | 5.00 | 3.4105 | 0.04795 | 184 | 71.13 | <.0001 |
| Treatment | 2 | 5.00 | 3.2971 | 0.04618 | 184 | 71.40 | <.0001 |
| Treatment | 3 | 5.00 | 3.2412 | 0.04623 | 185 | 70.10 | <.0001 |
| Treatment | 1 | 10.00 | 3.5043 | 0.04493 | 155 | 78.00 | <.0001 |
| Treatment | 2 | 10.00 | 3.4099 | 0.04334 | 156 | 78.67 | <.0001 |
| Treatment | 3 | 10.00 | 3.3032 | 0.04337 | 157 | 76.16 | <.0001 |
| Treatment | 1 | 15.00 | 3.4946 | 0.05014 | 204 | 69.70 | <.0001 |
| Treatment | 2 | 15.00 | 3.4047 | 0.04858 | 208 | 70.08 | <.0001 |
| Treatment | 3 | 15.00 | 3.2383 | 0.04877 | 210 | 66.40 | <.0001 |
| Treatment | 1 | 19.00 | 3.5710 | 0.07927 | 472 | 45.05 | <.0001 |
| Treatment | 2 | 19.00 | 3.3739 | 0.07679 | 470 | 43.94 | <.0001 |
| Treatment | 3 | 19.00 | 3.2264 | 0.07789 | 467 | 41.42 | <.0001 |
| Differences of Least Squares Means | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Effect | Treatment | _Treatment | Week | Estimate | Standard Error | DF | t Value | Pr > |t| | Adjustment | Adj P |
| Treatment | 1 | 2 | 1.00 | 0.03842 | 0.08464 | 375 | 0.45 | 0.6502 | Tukey-Kramer | 0.8927 |
| Treatment | 1 | 3 | 1.00 | 0.1329 | 0.08464 | 375 | 1.57 | 0.1171 | Tukey-Kramer | 0.2593 |
| Treatment | 2 | 3 | 1.00 | 0.09452 | 0.08299 | 375 | 1.14 | 0.2555 | Tukey-Kramer | 0.4905 |
| Treatment | 1 | 2 | 5.00 | 0.1134 | 0.06560 | 175 | 1.73 | 0.0856 | Tukey-Kramer | 0.1954 |
| Treatment | 1 | 3 | 5.00 | 0.1693 | 0.06565 | 176 | 2.58 | 0.0107 | Tukey-Kramer | 0.0274 |
| Treatment | 2 | 3 | 5.00 | 0.05585 | 0.06437 | 176 | 0.87 | 0.3868 | Tukey-Kramer | 0.6610 |
| Treatment | 1 | 2 | 10.00 | 0.09449 | 0.05998 | 139 | 1.58 | 0.1174 | Tukey-Kramer | 0.2571 |
| Treatment | 1 | 3 | 10.00 | 0.2012 | 0.06000 | 139 | 3.35 | 0.0010 | Tukey-Kramer | 0.0025 |
| Treatment | 2 | 3 | 10.00 | 0.1067 | 0.05884 | 139 | 1.81 | 0.0720 | Tukey-Kramer | 0.1662 |
| Treatment | 1 | 2 | 15.00 | 0.08995 | 0.06792 | 189 | 1.32 | 0.1870 | Tukey-Kramer | 0.3822 |
| Treatment | 1 | 3 | 15.00 | 0.2563 | 0.06803 | 190 | 3.77 | 0.0002 | Tukey-Kramer | 0.0005 |
| Treatment | 2 | 3 | 15.00 | 0.1663 | 0.06685 | 191 | 2.49 | 0.0137 | Tukey-Kramer | 0.0350 |
| Treatment | 1 | 2 | 19.00 | 0.1971 | 0.1091 | 440 | 1.81 | 0.0715 | Tukey-Kramer | 0.1682 |
| Treatment | 1 | 3 | 19.00 | 0.3446 | 0.1099 | 440 | 3.13 | 0.0018 | Tukey-Kramer | 0.0051 |
| Treatment | 2 | 3 | 19.00 | 0.1474 | 0.1082 | 438 | 1.36 | 0.1736 | Tukey-Kramer | 0.3612 |