Original Program from program editor.
*******************************************************************;
*** Neter, Kutner, Nachtsheim, Wasserman (1996) [CH19PR18.sas] ***;
*** Kidney failure patients on dialysis. Amount of dialysis ***;
*** needed depends on duration of treatment and weight gain ***;
*** from fluid retention In this study the effect of these two ***;
*** facters was to be evaluated by examining the number of ***;
*** days of hospitalization during the year. ***;
*******************************************************************;
options ps=256 ls=99 nocenter nodate nonumber nolabel;
TITLE1 'Factorial ANOVA (CRD with factorial treatment arrangement)';
data kidney; infile cards missover;
TITLE2 'Analysis of duration of patients stay in hospital';
LABEL days = 'Days in hospital during year';
LABEL duration = 'Duration of disease';
LABEL wtgain = 'Weight gain between visits (fluid retention)';
LABEL rep = 'Patient';
input days d w rep;
wtgain = 'Moderate';
if w = 1 then wtgain = 'Mild';
if w = 3 then wtgain = 'Severe';
duration = 'Short';
if d = 2 then duration = 'Long';
cards; run;
0.0 1 1 1
2.0 1 1 2
1.0 1 1 3
3.0 1 1 4
0.0 1 1 5
2.0 1 1 6
0.0 1 1 7
5.0 1 1 8
6.0 1 1 9
8.0 1 1 10
2.0 1 2 1
4.0 1 2 2
7.0 1 2 3
12.0 1 2 4
15.0 1 2 5
4.0 1 2 6
3.0 1 2 7
1.0 1 2 8
5.0 1 2 9
20.0 1 2 10
15.0 1 3 1
10.0 1 3 2
8.0 1 3 3
5.0 1 3 4
25.0 1 3 5
16.0 1 3 6
7.0 1 3 7
30.0 1 3 8
3.0 1 3 9
27.0 1 3 10
0.0 2 1 1
1.0 2 1 2
1.0 2 1 3
0.0 2 1 4
4.0 2 1 5
2.0 2 1 6
7.0 2 1 7
4.0 2 1 8
0.0 2 1 9
3.0 2 1 10
5.0 2 2 1
3.0 2 2 2
2.0 2 2 3
0.0 2 2 4
1.0 2 2 5
1.0 2 2 6
3.0 2 2 7
6.0 2 2 8
7.0 2 2 9
9.0 2 2 10
10.0 2 3 1
8.0 2 3 2
12.0 2 3 3
3.0 2 3 4
7.0 2 3 5
15.0 2 3 6
4.0 2 3 7
9.0 2 3 8
6.0 2 3 9
1.0 2 3 10
;
PROC PRINT DATA=kidney; TITLE3 'LISTING OF DATA'; RUN;
PROC MIXED DATA=kidney; CLASS WTGAIN DURATION;
TITLE3 'Analysis of Variance done with PROC MIXED';
MODEL DAYS = WTGAIN DURATION WTGAIN*DURATION / HType=3 ddfm=satterth outp=resids;
LSMEANS WTGAIN DURATION WTGAIN*DURATION / PDIFF ADJUST=TUKEY;
ods output diffs=ppp lsmeans=mmm;
**ods listing exclude diffs lsmeans; *this is now just a comment;
run;
TITLE4 'Post hoc adjustment with macro by Arnold Saxton';
* SAS Macro by Arnold Saxton: Saxton, A.M. 1998. A macro for ;
* converting mean separation output to letter groupings in Proc Mixed. ;
* In Proc. 23rd SAS Users Group Intl., SAS Institute, Cary, NC, pp1243-1246.;
%include 'C:\Geaghan\EXST\EXST7005New\Fall2003\SaS\pdmix800.sas';
%pdmix800(ppp,mmm,alpha=.01,sort=yes);
RUN;
QUIT;
proc univariate data=resids normal plot; var resid;
TITLE4 'Univariate analysis of RESIDUALS';
run;
GOPTIONS DEVICE=CGMflwa GSFMODE=REPLACE GSFNAME=OUT NOPROMPT noROTATE
ftext='TimesRoman' ftitle='TimesRoman';
FILENAME OUT 'C:\Geaghan\EXST\EXST7005New\Fall2003\_Disk_Fall03\hospital.CGM';
PROC GCHART data=kidney; title3 'WTGAIN by DURATION';
pattern v=s color=black;
block WTGAIN / group=DURATION discrete type=mean sumvar=DAYS;
run; quit;
PROC GLM DATA=kidney; CLASS WTGAIN DURATION;
TITLE3 'Example of a 2 way ANOVA done with PROC GLM';
MODEL DAYS = WTGAIN DURATION WTGAIN*DURATION / SS3;
LSMEANS WTGAIN DURATION WTGAIN*DURATION / PDIFF STDERR ADJUST=TUKEY;
output out=next residual=e;
RUN; QUIT;
Below is output from the SAS log (bold) and output from the SAS
Output window.
1 *******************************************************************;
2 *** Neter, Kutner, Nachtsheim, Wasserman (1996) [CH19PR18.sas] ***;
3 *** Kidney failure patients on dialysis. Amount of dialysis ***;
4 *** needed depends on duration of treatment and weight gain ***;
5 *** from fluid retention In this study the effect of these two ***;
6 *** facters was to be evaluated by examining the number of ***;
7 *** days of hospitalization during the year. ***;
8 *******************************************************************;
9 options ps=256 ls=99 nocenter nodate nonumber nolabel;
10 TITLE1 'Factorial ANOVA (CRD with factorial treatment arrangement)';
11
12
13 data kidney; infile cards missover;
14 TITLE2 'Analysis of duration of patients stay in hospital';
15 LABEL days = 'Days in hospital during year';
16 LABEL duration = 'Duration of disease';
17 LABEL wtgain = 'Weight gain between visits (fluid retention)';
18 LABEL rep = 'Patient';
19 input days d w rep;
20 wtgain = 'Moderate';
21 if w = 1 then wtgain = 'Mild';
22 if w = 3 then wtgain = 'Severe';
23 duration = 'Short';
24 if d = 2 then duration = 'Long';
25 cards;
NOTE: The data set WORK.KIDNEY has 60 observations and 6 variables.
NOTE: DATA statement used (Total process time):
real time 0.03 seconds
cpu time 0.03 seconds
25 ! run;
86 ;
87 PROC PRINT DATA=kidney; TITLE3 'LISTING OF DATA'; RUN;
NOTE: There were 60 observations read from the data set WORK.KIDNEY.
NOTE: The PROCEDURE PRINT printed page 1.
NOTE: PROCEDURE PRINT used (Total process time):
real time 0.00 seconds
cpu time 0.00 seconds
Factorial ANOVA (CRD with factorial treatment arrangement)
Analysis of duration of patients stay in hospital
LISTING OF DATA
Obs days duration wtgain rep d w
1 0 Short Mild 1 1 1
2 2 Short Mild 2 1 1
3 1 Short Mild 3 1 1
4 3 Short Mild 4 1 1
5 0 Short Mild 5 1 1
6 2 Short Mild 6 1 1
7 0 Short Mild 7 1 1
8 5 Short Mild 8 1 1
9 6 Short Mild 9 1 1
10 8 Short Mild 10 1 1
11 2 Short Moderate 1 1 2
12 4 Short Moderate 2 1 2
13 7 Short Moderate 3 1 2
14 12 Short Moderate 4 1 2
15 15 Short Moderate 5 1 2
16 4 Short Moderate 6 1 2
17 3 Short Moderate 7 1 2
18 1 Short Moderate 8 1 2
19 5 Short Moderate 9 1 2
20 20 Short Moderate 10 1 2
21 15 Short Severe 1 1 3
22 10 Short Severe 2 1 3
23 8 Short Severe 3 1 3
24 5 Short Severe 4 1 3
25 25 Short Severe 5 1 3
26 16 Short Severe 6 1 3
27 7 Short Severe 7 1 3
28 30 Short Severe 8 1 3
29 3 Short Severe 9 1 3
30 27 Short Severe 10 1 3
31 0 Long Mild 1 2 1
32 1 Long Mild 2 2 1
33 1 Long Mild 3 2 1
34 0 Long Mild 4 2 1
35 4 Long Mild 5 2 1
36 2 Long Mild 6 2 1
37 7 Long Mild 7 2 1
38 4 Long Mild 8 2 1
39 0 Long Mild 9 2 1
40 3 Long Mild 10 2 1
41 5 Long Moderate 1 2 2
42 3 Long Moderate 2 2 2
43 2 Long Moderate 3 2 2
44 0 Long Moderate 4 2 2
45 1 Long Moderate 5 2 2
46 1 Long Moderate 6 2 2
47 3 Long Moderate 7 2 2
48 6 Long Moderate 8 2 2
49 7 Long Moderate 9 2 2
50 9 Long Moderate 10 2 2
51 10 Long Severe 1 2 3
52 8 Long Severe 2 2 3
53 12 Long Severe 3 2 3
54 3 Long Severe 4 2 3
55 7 Long Severe 5 2 3
56 15 Long Severe 6 2 3
57 4 Long Severe 7 2 3
58 9 Long Severe 8 2 3
59 6 Long Severe 9 2 3
60 1 Long Severe 10 2 3
88
89 PROC MIXED DATA=kidney; CLASS WTGAIN DURATION;
90 TITLE3 'Analysis of Variance done with PROC MIXED';
91 MODEL DAYS = WTGAIN DURATION WTGAIN*DURATION / HType=3 ddfm=satterth outp=resids;
92 LSMEANS WTGAIN DURATION WTGAIN*DURATION / PDIFF ADJUST=TUKEY;
93 ods output diffs=ppp lsmeans=mmm;
94 **ods listing exclude diffs lsmeans; *this is now just a comment;
95 run;
NOTE: The data set WORK.MMM has 11 observations and 8 variables.
NOTE: The data set WORK.PPP has 19 observations and 12 variables.
NOTE: The data set WORK.RESIDS has 60 observations and 13 variables.
NOTE: The PROCEDURE MIXED printed page 2.
NOTE: PROCEDURE MIXED used (Total process time):
real time 0.09 seconds
cpu time 0.07 seconds
96 TITLE4 'Post hoc adjustment with macro by Arnold Saxton';
97 * SAS Macro by Arnold Saxton: Saxton, A.M. 1998. A macro for ;
98 * converting mean separation output to letter groupings in Proc Mixed. ;
99 * In Proc. 23rd SAS Users Group Intl., SAS Institute, Cary, NC, pp1243-1246.;
100 %include 'C:\Geaghan\EXST\EXST7005New\Fall2003\SaS\pdmix800.sas';
728 %pdmix800(ppp,mmm,alpha=.01,sort=yes);
PDMIX800 03.26.2002 processing
4.3008418397
Tukey values for wtgain are 5.17691 (avg) 5.17691 (min) 5.17691 (max).
3.7759475003
Tukey values for duration are 3.71106 (avg) 3.71106 (min) 3.71106 (max).
5.018342024
Tukey values for wtgain*duration are 8.54265 (avg) 8.54265 (min) 8.54265 (max).
729 RUN;
730 QUIT;
Factorial ANOVA (CRD with factorial treatment arrangement)
Analysis of duration of patients stay in hospital
Analysis of Variance done with PROC MIXED
The Mixed Procedure
Model Information
Data Set WORK.KIDNEY
Dependent Variable days
Covariance Structure Diagonal
Estimation Method REML
Residual Variance Method Profile
Fixed Effects SE Method Model-Based
Degrees of Freedom Method Residual
Class Level Information
Class Levels Values
wtgain 3 Mild Moderate Severe
duration 2 Long Short
Dimensions
Covariance Parameters 1
Columns in X 12
Columns in Z 0
Subjects 1
Max Obs Per Subject 60
Observations Used 60
Observations Not Used 0
Total Observations 60
Covariance Parameter Estimates
Cov Parm Estimate
Residual 28.9778
Fit Statistics
-2 Res Log Likelihood 348.9
AIC (smaller is better) 350.9
AICC (smaller is better) 350.9
BIC (smaller is better) 352.8
Type 3 Tests of Fixed Effects
Num Den
Effect DF DF F Value Pr > F
wtgain 2 54 13.12 <.0001
duration 1 54 7.21 0.0096
wtgain*duration 2 54 1.88 0.1622
Least Squares Means
Standard
Effect wtgain duration Estimate Error DF t Value Pr > |t|
wtgain Mild 2.4500 1.2037 54 2.04 0.0467
wtgain Moderate 5.5000 1.2037 54 4.57 <.0001
wtgain Severe 11.0500 1.2037 54 9.18 <.0001
duration Long 4.4667 0.9828 54 4.54 <.0001
duration Short 8.2000 0.9828 54 8.34 <.0001
wtgain*duration Mild Long 2.2000 1.7023 54 1.29 0.2017
wtgain*duration Mild Short 2.7000 1.7023 54 1.59 0.1186
wtgain*duration Moderate Long 3.7000 1.7023 54 2.17 0.0341
wtgain*duration Moderate Short 7.3000 1.7023 54 4.29 <.0001
wtgain*duration Severe Long 7.5000 1.7023 54 4.41 <.0001
wtgain*duration Severe Short 14.6000 1.7023 54 8.58 <.0001
Differences ofLeast Squares Means
Standard
Effect wtgain duration _wtgain _duration Estimate Error DF t Value
wtgain Mild Moderate -3.0500 1.7023 54 -1.79
wtgain Mild Severe -8.6000 1.7023 54 -5.05
wtgain Moderate Severe -5.5500 1.7023 54 -3.26
duration Long Short -3.7333 1.3899 54 -2.69
wtgain*duration Mild Long Mild Short -0.5000 2.4074 54 -0.21
wtgain*duration Mild Long Moderate Long -1.5000 2.4074 54 -0.62
wtgain*duration Mild Long Moderate Short -5.1000 2.4074 54 -2.12
wtgain*duration Mild Long Severe Long -5.3000 2.4074 54 -2.20
wtgain*duration Mild Long Severe Short -12.4000 2.4074 54 -5.15
wtgain*duration Mild Short Moderate Long -1.0000 2.4074 54 -0.42
wtgain*duration Mild Short Moderate Short -4.6000 2.4074 54 -1.91
wtgain*duration Mild Short Severe Long -4.8000 2.4074 54 -1.99
wtgain*duration Mild Short Severe Short -11.9000 2.4074 54 -4.94
wtgain*duration Moderate Long Moderate Short -3.6000 2.4074 54 -1.50
wtgain*duration Moderate Long Severe Long -3.8000 2.4074 54 -1.58
wtgain*duration Moderate Long Severe Short -10.9000 2.4074 54 -4.53
wtgain*duration Moderate Short Severe Long -0.2000 2.4074 54 -0.08
wtgain*duration Moderate Short Severe Short -7.3000 2.4074 54 -3.03
wtgain*duration Severe Long Severe Short -7.1000 2.4074 54 -2.95
Differences ofLeast Squares Means
Effect wtgain duration _wtgain _duration Pr > |t| Adjustment Adj P
wtgain Mild Moderate 0.0788 Tukey 0.1820
wtgain Mild Severe <.0001 Tukey <.0001
wtgain Moderate Severe 0.0019 Tukey 0.0054
duration Long Short 0.0096 Tukey 0.0096
wtgain*duration Mild Long Mild Short 0.8362 Tukey 0.9999
wtgain*duration Mild Long Moderate Long 0.5359 Tukey 0.9888
wtgain*duration Mild Long Moderate Short 0.0388 Tukey 0.2935
wtgain*duration Mild Long Severe Long 0.0320 Tukey 0.2540
wtgain*duration Mild Long Severe Short <.0001 Tukey <.0001
wtgain*duration Mild Short Moderate Long 0.6795 Tukey 0.9983
wtgain*duration Mild Short Moderate Short 0.0613 Tukey 0.4069
wtgain*duration Mild Short Severe Long 0.0512 Tukey 0.3592
wtgain*duration Mild Short Severe Short <.0001 Tukey 0.0001
wtgain*duration Moderate Long Moderate Short 0.1406 Tukey 0.6686
wtgain*duration Moderate Long Severe Long 0.1203 Tukey 0.6159
wtgain*duration Moderate Long Severe Short <.0001 Tukey 0.0005
wtgain*duration Moderate Short Severe Long 0.9341 Tukey 1.0000
wtgain*duration Moderate Short Severe Short 0.0037 Tukey 0.0411
wtgain*duration Severe Long Severe Short 0.0047 Tukey 0.0507
Factorial ANOVA (CRD with factorial treatment arrangement)
Analysis of duration of patients stay in hospital
Analysis of Variance done with PROC MIXED
Post hoc adjustment with macro by Arnold Saxton
Effect=wtgain ADJUSTMENT=Tukey(P<.01) BYGROUP=1
Obs wtgain duration Estimate StdErr MSGROUP
1 Severe 11.0500 1.2037 A
2 Moderate 5.5000 1.2037 B
3 Mild 2.4500 1.2037 B
Effect=duration ADJUSTMENT=Tukey(P<.01) BYGROUP=2
Obs wtgain duration Estimate StdErr MSGROUP
4 Short 8.2000 0.9828 A
5 Long 4.4667 0.9828 B
Effect=wtgain*duration ADJUSTMENT=Tukey(P<.01) BYGROUP=3
Obs wtgain duration Estimate StdErr MSGROUP
6 Severe Short 14.6000 1.7023 A
7 Severe Long 7.5000 1.7023 AB
8 Moderate Short 7.3000 1.7023 AB
9 Moderate Long 3.7000 1.7023 B
10 Mild Short 2.7000 1.7023 B
11 Mild Long 2.2000 1.7023 B
731
732 proc univariate data=resids normal plot; var resid;
733 TITLE4 'Univariate analysis of RESIDUALS';
734 run;
NOTE: The PROCEDURE UNIVARIATE printed page 4.
NOTE: PROCEDURE UNIVARIATE used (Total process time):
real time 0.01 seconds
cpu time 0.01 seconds
Factorial ANOVA (CRD with factorial treatment arrangement)
Analysis of duration of patients stay in hospital
Analysis of Variance done with PROC MIXED
Univariate analysis of RESIDUALS
The UNIVARIATE Procedure
Variable: Resid
Moments
N 60 Sum Weights 60
Mean 0 Sum Observations 0
Std Deviation 5.14995475 Variance 26.5220339
Skewness 0.75823095 Kurtosis 1.26399843
Uncorrected SS 1564.8 Corrected SS 1564.8
Coeff Variation . Std Error Mean 0.6648563
Basic Statistical Measures
Location Variability
Mean 0.00000 Std Deviation 5.14995
Median -0.70000 Variance 26.52203
Mode -2.70000 Range 27.00000
Interquartile Range 5.00000
NOTE: The mode displayed is the smallest of 2 modes with a count of 3.
Tests for Location: Mu0=0n
Test -Statistic- -----p Value------
Student's t t 0 Pr > |t| 1.0000
Sign M -5 Pr >= |M| 0.2451
Signed Rank S -89 Pr >= |S| 0.5169
Tests for Normality
Test --Statistic--- -----p Value------
Shapiro-Wilk W 0.954062 Pr < W 0.0243
Kolmogorov-Smirnov D 0.098822 Pr > D 0.1496
Cramer-von Mises W-Sq 0.162545 Pr > W-Sq 0.0171
Anderson-Darling A-Sq 0.96946 Pr > A-Sq 0.0149
Quantiles (Definition 5)
Quantile Estimate
100% Max 15.4
99% 15.4
95% 11.4
90% 6.4
75% Q3 2.3
50% Median -0.7
25% Q1 -2.7
10% -5.8
5% -7.1
1% -11.6
0% Min -11.6
Extreme Observations
----Lowest---- ----Highest---
Value Obs Value Obs
-11.6 29 7.7 15
-9.6 24 10.4 25
-7.6 27 12.4 30
-6.6 23 12.7 20
-6.5 60 15.4 28
Stem Leaf Boxplot
14 4 1 0
12 47 2 0
10 4 1 0
8
6 57 2 |
4 57833 5 |
2 33533 5 +-----+
0 345834588 9 | + |
-0 775227777532 12 *-----*
-2 7533777773222 13 +-----+
-4 3653 4 |
-6 6653 4 |
-8 6 1 |
-10 6 1 0
----+----+----+----+
Normal Probability Plot
15+ *
| * * ++
| * ++++
| ++++
| +**+
| +****
| ++****
| ++****
| ******
| ******
| ***++
| **+**+
| *+++
-11+ *+++
+----+----+----+----+----+----+----+----+----+----+
-2 -1 0 +1 +2
737 GOPTIONS DEVICE=CGMflwa GSFMODE=REPLACE GSFNAME=OUT NOPROMPT noROTATE
738 ftext='TimesRoman' ftitle='TimesRoman';
739 FILENAME OUT 'C:\Geaghan\EXST\EXST7005New\Fall2003\_Disk_Fall03\hospital.CGM';
740 PROC GCHART data=kidney; title3 'WTGAIN by DURATION';
741 pattern v=s color=black;
742 block WTGAIN / group=DURATION discrete type=mean sumvar=DAYS;
743 run;
WARNING: TITLE1 is too long. Height has been reduced to 86.21 pct of specified or default size.
NOTE: Foreground color BLACK same as background. Part of your graph may not be visible.
NOTE: 9 RECORDS WRITTEN TO C:\Geaghan\EXST\EXST7005New\Fall2003\_Disk_Fall03\hospital.CGM
743 ! quit;
NOTE: There were 60 observations read from the data set WORK.KIDNEY.
NOTE: PROCEDURE GCHART used (Total process time):
real time 0.07 seconds
cpu time 0.01 seconds
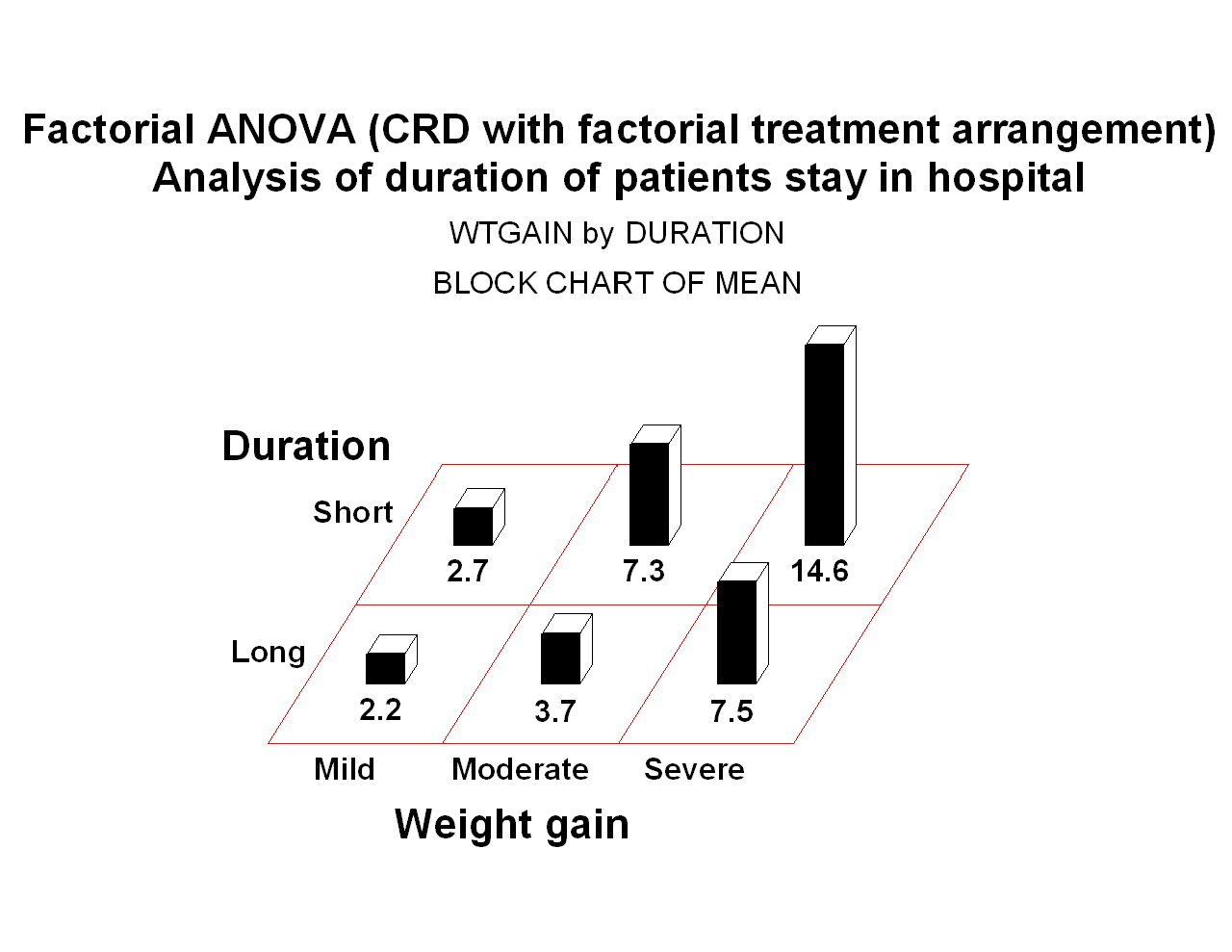 745
746 PROC GLM DATA=kidney; CLASS WTGAIN DURATION;
747 TITLE3 'Example of a 2 way ANOVA done with PROC GLM';
748 MODEL DAYS = WTGAIN DURATION WTGAIN*DURATION / SS3;
749 LSMEANS WTGAIN DURATION WTGAIN*DURATION / PDIFF STDERR ADJUST=TUKEY;
750 output out=next residual=e;
751 RUN;
751 ! QUIT;
NOTE: The data set WORK.NEXT has 60 observations and 7 variables.
NOTE: The PROCEDURE GLM printed pages 5-9.
NOTE: PROCEDURE GLM used (Total process time):
real time 0.07 seconds
cpu time 0.07 seconds
NOTE: SAS Institute Inc., SAS Campus Drive, Cary, NC USA 27513-2414
NOTE: The SAS System used:
real time 1.73 seconds
cpu time 0.87 seconds
Factorial ANOVA (CRD with factorial treatment arrangement)
Analysis of duration of patients stay in hospital
Example of a 2 way ANOVA done with PROC GLM
The GLM Procedure
Class Level Information
Class Levels Values
wtgain 3 Mild Moderate Severe
duration 2 Long Short
Number of observations 60
Dependent Variable: days
Sum of
Source DF Squares Mean Square F Value Pr > F
Model 5 1078.533333 215.706667 7.44 <.0001
Error 54 1564.800000 28.977778
Corrected Total 59 2643.333333
R-Square Coeff Var Root MSE days Mean
0.408020 84.99633 5.383101 6.333333
Source DF Type III SS Mean Square F Value Pr > F
wtgain 2 760.4333333 380.2166667 13.12 <.0001
duration 1 209.0666667 209.0666667 7.21 0.0096
wtgain*duration 2 109.0333333 54.5166667 1.88 0.1622
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
Standard LSMEAN
wtgain days LSMEAN Error Pr > |t| Number
Mild 2.4500000 1.2036980 0.0467 1
Moderate 5.5000000 1.2036980 <.0001 2
Severe 11.0500000 1.2036980 <.0001 3
Least Squares Means for effect wtgain
Pr > |t| for H0: LSMean(i)=LSMean(j)
Dependent Variable: days
i/j 1 2 3
1 0.1820 <.0001
2 0.1820 0.0054
3 <.0001 0.0054
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
H0:LSMean1=
Standard H0:LSMEAN=0 LSMean2
duration days LSMEAN Error Pr > |t| Pr > |t|
Long 4.46666667 0.98281531 <.0001 0.0096
Short 8.20000000 0.98281531 <.0001
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
Standard LSMEAN
wtgain duration days LSMEAN Error Pr > |t| Number
Mild Long 2.2000000 1.7022860 0.2017 1
Mild Short 2.7000000 1.7022860 0.1186 2
Moderate Long 3.7000000 1.7022860 0.0341 3
Moderate Short 7.3000000 1.7022860 <.0001 4
Severe Long 7.5000000 1.7022860 <.0001 5
Severe Short 14.6000000 1.7022860 <.0001 6
Least Squares Means for effect wtgain*duration
Pr > |t| for H0: LSMean(i)=LSMean(j)
Dependent Variable: days
i/j 1 2 3 4 5 6
1 0.9999 0.9888 0.2935 0.2540 <.0001
2 0.9999 0.9983 0.4069 0.3592 0.0001
3 0.9888 0.9983 0.6686 0.6159 0.0005
4 0.2935 0.4069 0.6686 1.0000 0.0411
5 0.2540 0.3592 0.6159 1.0000 0.0507
6 <.0001 0.0001 0.0005 0.0411 0.0507
745
746 PROC GLM DATA=kidney; CLASS WTGAIN DURATION;
747 TITLE3 'Example of a 2 way ANOVA done with PROC GLM';
748 MODEL DAYS = WTGAIN DURATION WTGAIN*DURATION / SS3;
749 LSMEANS WTGAIN DURATION WTGAIN*DURATION / PDIFF STDERR ADJUST=TUKEY;
750 output out=next residual=e;
751 RUN;
751 ! QUIT;
NOTE: The data set WORK.NEXT has 60 observations and 7 variables.
NOTE: The PROCEDURE GLM printed pages 5-9.
NOTE: PROCEDURE GLM used (Total process time):
real time 0.07 seconds
cpu time 0.07 seconds
NOTE: SAS Institute Inc., SAS Campus Drive, Cary, NC USA 27513-2414
NOTE: The SAS System used:
real time 1.73 seconds
cpu time 0.87 seconds
Factorial ANOVA (CRD with factorial treatment arrangement)
Analysis of duration of patients stay in hospital
Example of a 2 way ANOVA done with PROC GLM
The GLM Procedure
Class Level Information
Class Levels Values
wtgain 3 Mild Moderate Severe
duration 2 Long Short
Number of observations 60
Dependent Variable: days
Sum of
Source DF Squares Mean Square F Value Pr > F
Model 5 1078.533333 215.706667 7.44 <.0001
Error 54 1564.800000 28.977778
Corrected Total 59 2643.333333
R-Square Coeff Var Root MSE days Mean
0.408020 84.99633 5.383101 6.333333
Source DF Type III SS Mean Square F Value Pr > F
wtgain 2 760.4333333 380.2166667 13.12 <.0001
duration 1 209.0666667 209.0666667 7.21 0.0096
wtgain*duration 2 109.0333333 54.5166667 1.88 0.1622
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
Standard LSMEAN
wtgain days LSMEAN Error Pr > |t| Number
Mild 2.4500000 1.2036980 0.0467 1
Moderate 5.5000000 1.2036980 <.0001 2
Severe 11.0500000 1.2036980 <.0001 3
Least Squares Means for effect wtgain
Pr > |t| for H0: LSMean(i)=LSMean(j)
Dependent Variable: days
i/j 1 2 3
1 0.1820 <.0001
2 0.1820 0.0054
3 <.0001 0.0054
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
H0:LSMean1=
Standard H0:LSMEAN=0 LSMean2
duration days LSMEAN Error Pr > |t| Pr > |t|
Long 4.46666667 0.98281531 <.0001 0.0096
Short 8.20000000 0.98281531 <.0001
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
Standard LSMEAN
wtgain duration days LSMEAN Error Pr > |t| Number
Mild Long 2.2000000 1.7022860 0.2017 1
Mild Short 2.7000000 1.7022860 0.1186 2
Moderate Long 3.7000000 1.7022860 0.0341 3
Moderate Short 7.3000000 1.7022860 <.0001 4
Severe Long 7.5000000 1.7022860 <.0001 5
Severe Short 14.6000000 1.7022860 <.0001 6
Least Squares Means for effect wtgain*duration
Pr > |t| for H0: LSMean(i)=LSMean(j)
Dependent Variable: days
i/j 1 2 3 4 5 6
1 0.9999 0.9888 0.2935 0.2540 <.0001
2 0.9999 0.9983 0.4069 0.3592 0.0001
3 0.9888 0.9983 0.6686 0.6159 0.0005
4 0.2935 0.4069 0.6686 1.0000 0.0411
5 0.2540 0.3592 0.6159 1.0000 0.0507
6 <.0001 0.0001 0.0005 0.0411 0.0507
Last modified
by James P. Geaghan
on Wednesday, August 13, 2003
 745
746 PROC GLM DATA=kidney; CLASS WTGAIN DURATION;
747 TITLE3 'Example of a 2 way ANOVA done with PROC GLM';
748 MODEL DAYS = WTGAIN DURATION WTGAIN*DURATION / SS3;
749 LSMEANS WTGAIN DURATION WTGAIN*DURATION / PDIFF STDERR ADJUST=TUKEY;
750 output out=next residual=e;
751 RUN;
751 ! QUIT;
NOTE: The data set WORK.NEXT has 60 observations and 7 variables.
NOTE: The PROCEDURE GLM printed pages 5-9.
NOTE: PROCEDURE GLM used (Total process time):
real time 0.07 seconds
cpu time 0.07 seconds
NOTE: SAS Institute Inc., SAS Campus Drive, Cary, NC USA 27513-2414
NOTE: The SAS System used:
real time 1.73 seconds
cpu time 0.87 seconds
Factorial ANOVA (CRD with factorial treatment arrangement)
Analysis of duration of patients stay in hospital
Example of a 2 way ANOVA done with PROC GLM
The GLM Procedure
Class Level Information
Class Levels Values
wtgain 3 Mild Moderate Severe
duration 2 Long Short
Number of observations 60
Dependent Variable: days
Sum of
Source DF Squares Mean Square F Value Pr > F
Model 5 1078.533333 215.706667 7.44 <.0001
Error 54 1564.800000 28.977778
Corrected Total 59 2643.333333
R-Square Coeff Var Root MSE days Mean
0.408020 84.99633 5.383101 6.333333
Source DF Type III SS Mean Square F Value Pr > F
wtgain 2 760.4333333 380.2166667 13.12 <.0001
duration 1 209.0666667 209.0666667 7.21 0.0096
wtgain*duration 2 109.0333333 54.5166667 1.88 0.1622
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
Standard LSMEAN
wtgain days LSMEAN Error Pr > |t| Number
Mild 2.4500000 1.2036980 0.0467 1
Moderate 5.5000000 1.2036980 <.0001 2
Severe 11.0500000 1.2036980 <.0001 3
Least Squares Means for effect wtgain
Pr > |t| for H0: LSMean(i)=LSMean(j)
Dependent Variable: days
i/j 1 2 3
1 0.1820 <.0001
2 0.1820 0.0054
3 <.0001 0.0054
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
H0:LSMean1=
Standard H0:LSMEAN=0 LSMean2
duration days LSMEAN Error Pr > |t| Pr > |t|
Long 4.46666667 0.98281531 <.0001 0.0096
Short 8.20000000 0.98281531 <.0001
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
Standard LSMEAN
wtgain duration days LSMEAN Error Pr > |t| Number
Mild Long 2.2000000 1.7022860 0.2017 1
Mild Short 2.7000000 1.7022860 0.1186 2
Moderate Long 3.7000000 1.7022860 0.0341 3
Moderate Short 7.3000000 1.7022860 <.0001 4
Severe Long 7.5000000 1.7022860 <.0001 5
Severe Short 14.6000000 1.7022860 <.0001 6
Least Squares Means for effect wtgain*duration
Pr > |t| for H0: LSMean(i)=LSMean(j)
Dependent Variable: days
i/j 1 2 3 4 5 6
1 0.9999 0.9888 0.2935 0.2540 <.0001
2 0.9999 0.9983 0.4069 0.3592 0.0001
3 0.9888 0.9983 0.6686 0.6159 0.0005
4 0.2935 0.4069 0.6686 1.0000 0.0411
5 0.2540 0.3592 0.6159 1.0000 0.0507
6 <.0001 0.0001 0.0005 0.0411 0.0507
745
746 PROC GLM DATA=kidney; CLASS WTGAIN DURATION;
747 TITLE3 'Example of a 2 way ANOVA done with PROC GLM';
748 MODEL DAYS = WTGAIN DURATION WTGAIN*DURATION / SS3;
749 LSMEANS WTGAIN DURATION WTGAIN*DURATION / PDIFF STDERR ADJUST=TUKEY;
750 output out=next residual=e;
751 RUN;
751 ! QUIT;
NOTE: The data set WORK.NEXT has 60 observations and 7 variables.
NOTE: The PROCEDURE GLM printed pages 5-9.
NOTE: PROCEDURE GLM used (Total process time):
real time 0.07 seconds
cpu time 0.07 seconds
NOTE: SAS Institute Inc., SAS Campus Drive, Cary, NC USA 27513-2414
NOTE: The SAS System used:
real time 1.73 seconds
cpu time 0.87 seconds
Factorial ANOVA (CRD with factorial treatment arrangement)
Analysis of duration of patients stay in hospital
Example of a 2 way ANOVA done with PROC GLM
The GLM Procedure
Class Level Information
Class Levels Values
wtgain 3 Mild Moderate Severe
duration 2 Long Short
Number of observations 60
Dependent Variable: days
Sum of
Source DF Squares Mean Square F Value Pr > F
Model 5 1078.533333 215.706667 7.44 <.0001
Error 54 1564.800000 28.977778
Corrected Total 59 2643.333333
R-Square Coeff Var Root MSE days Mean
0.408020 84.99633 5.383101 6.333333
Source DF Type III SS Mean Square F Value Pr > F
wtgain 2 760.4333333 380.2166667 13.12 <.0001
duration 1 209.0666667 209.0666667 7.21 0.0096
wtgain*duration 2 109.0333333 54.5166667 1.88 0.1622
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
Standard LSMEAN
wtgain days LSMEAN Error Pr > |t| Number
Mild 2.4500000 1.2036980 0.0467 1
Moderate 5.5000000 1.2036980 <.0001 2
Severe 11.0500000 1.2036980 <.0001 3
Least Squares Means for effect wtgain
Pr > |t| for H0: LSMean(i)=LSMean(j)
Dependent Variable: days
i/j 1 2 3
1 0.1820 <.0001
2 0.1820 0.0054
3 <.0001 0.0054
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
H0:LSMean1=
Standard H0:LSMEAN=0 LSMean2
duration days LSMEAN Error Pr > |t| Pr > |t|
Long 4.46666667 0.98281531 <.0001 0.0096
Short 8.20000000 0.98281531 <.0001
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
Standard LSMEAN
wtgain duration days LSMEAN Error Pr > |t| Number
Mild Long 2.2000000 1.7022860 0.2017 1
Mild Short 2.7000000 1.7022860 0.1186 2
Moderate Long 3.7000000 1.7022860 0.0341 3
Moderate Short 7.3000000 1.7022860 <.0001 4
Severe Long 7.5000000 1.7022860 <.0001 5
Severe Short 14.6000000 1.7022860 <.0001 6
Least Squares Means for effect wtgain*duration
Pr > |t| for H0: LSMean(i)=LSMean(j)
Dependent Variable: days
i/j 1 2 3 4 5 6
1 0.9999 0.9888 0.2935 0.2540 <.0001
2 0.9999 0.9983 0.4069 0.3592 0.0001
3 0.9888 0.9983 0.6686 0.6159 0.0005
4 0.2935 0.4069 0.6686 1.0000 0.0411
5 0.2540 0.3592 0.6159 1.0000 0.0507
6 <.0001 0.0001 0.0005 0.0411 0.0507